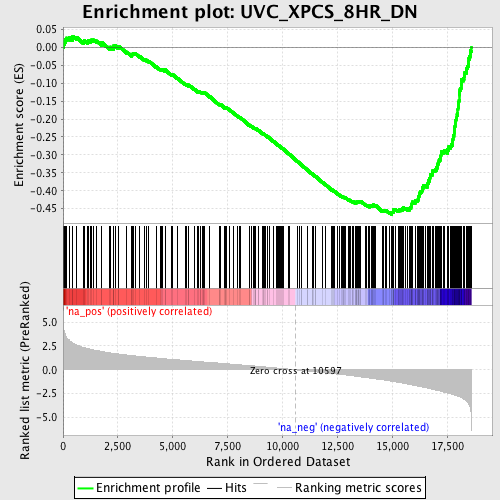
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | UVC_XPCS_8HR_DN |
| Enrichment Score (ES) | -0.46624723 |
| Normalized Enrichment Score (NES) | -2.4986215 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CRIM1 | 22 | 4.529 | 0.0091 | No | ||
| 2 | GNAQ | 64 | 3.955 | 0.0159 | No | ||
| 3 | DLC1 | 109 | 3.570 | 0.0216 | No | ||
| 4 | NUP98 | 169 | 3.340 | 0.0260 | No | ||
| 5 | ABCE1 | 279 | 3.086 | 0.0271 | No | ||
| 6 | RB1CC1 | 421 | 2.822 | 0.0258 | No | ||
| 7 | PMM2 | 440 | 2.798 | 0.0312 | No | ||
| 8 | LHFPL2 | 613 | 2.599 | 0.0278 | No | ||
| 9 | TRIM2 | 943 | 2.323 | 0.0151 | No | ||
| 10 | IGF2BP3 | 965 | 2.304 | 0.0192 | No | ||
| 11 | ARHGEF10 | 1119 | 2.212 | 0.0159 | No | ||
| 12 | EGFR | 1159 | 2.187 | 0.0188 | No | ||
| 13 | DOCK4 | 1257 | 2.133 | 0.0183 | No | ||
| 14 | ZHX3 | 1305 | 2.107 | 0.0206 | No | ||
| 15 | NRG1 | 1392 | 2.072 | 0.0206 | No | ||
| 16 | FYN | 1528 | 2.012 | 0.0178 | No | ||
| 17 | ATRX | 1753 | 1.921 | 0.0100 | No | ||
| 18 | RNF13 | 1757 | 1.920 | 0.0142 | No | ||
| 19 | EXT1 | 2128 | 1.776 | -0.0020 | No | ||
| 20 | IGF1R | 2150 | 1.768 | 0.0009 | No | ||
| 21 | CBLB | 2285 | 1.721 | -0.0025 | No | ||
| 22 | FGF2 | 2313 | 1.711 | -0.0000 | No | ||
| 23 | SMAD3 | 2315 | 1.711 | 0.0038 | No | ||
| 24 | PAWR | 2373 | 1.694 | 0.0046 | No | ||
| 25 | PKP4 | 2513 | 1.659 | 0.0008 | No | ||
| 26 | PTX3 | 2544 | 1.650 | 0.0029 | No | ||
| 27 | USP24 | 2900 | 1.545 | -0.0130 | No | ||
| 28 | CENPE | 3121 | 1.491 | -0.0216 | No | ||
| 29 | TLE1 | 3135 | 1.487 | -0.0189 | No | ||
| 30 | MGLL | 3184 | 1.472 | -0.0182 | No | ||
| 31 | SRGAP2 | 3213 | 1.466 | -0.0164 | No | ||
| 32 | ANKS1A | 3286 | 1.445 | -0.0170 | No | ||
| 33 | NR2F2 | 3482 | 1.401 | -0.0244 | No | ||
| 34 | WASF3 | 3704 | 1.350 | -0.0334 | No | ||
| 35 | HOMER1 | 3796 | 1.330 | -0.0354 | No | ||
| 36 | MYST3 | 3903 | 1.304 | -0.0382 | No | ||
| 37 | RYK | 4240 | 1.238 | -0.0537 | No | ||
| 38 | ARHGEF7 | 4422 | 1.194 | -0.0608 | No | ||
| 39 | TRAM2 | 4467 | 1.185 | -0.0605 | No | ||
| 40 | INPP5F | 4542 | 1.166 | -0.0619 | No | ||
| 41 | CUL2 | 4647 | 1.147 | -0.0650 | No | ||
| 42 | SERPINE1 | 4654 | 1.146 | -0.0627 | No | ||
| 43 | TIAM1 | 4950 | 1.089 | -0.0763 | No | ||
| 44 | PRKCI | 4989 | 1.083 | -0.0759 | No | ||
| 45 | NEDD4 | 5216 | 1.039 | -0.0859 | No | ||
| 46 | CXCL12 | 5563 | 0.972 | -0.1025 | No | ||
| 47 | OSBPL8 | 5626 | 0.960 | -0.1037 | No | ||
| 48 | RAGE | 5721 | 0.944 | -0.1067 | No | ||
| 49 | DOCK9 | 5727 | 0.943 | -0.1048 | No | ||
| 50 | CREB3L2 | 5980 | 0.896 | -0.1165 | No | ||
| 51 | RGS7 | 6140 | 0.866 | -0.1232 | No | ||
| 52 | MYO1B | 6184 | 0.857 | -0.1236 | No | ||
| 53 | PARN | 6245 | 0.847 | -0.1250 | No | ||
| 54 | CD2AP | 6276 | 0.840 | -0.1247 | No | ||
| 55 | PPP2R5C | 6354 | 0.829 | -0.1270 | No | ||
| 56 | TGFB2 | 6387 | 0.822 | -0.1269 | No | ||
| 57 | ZCCHC11 | 6410 | 0.817 | -0.1262 | No | ||
| 58 | HOXB2 | 6450 | 0.810 | -0.1265 | No | ||
| 59 | TSPAN5 | 6687 | 0.766 | -0.1376 | No | ||
| 60 | SLC7A1 | 7114 | 0.696 | -0.1593 | No | ||
| 61 | KIF11 | 7145 | 0.690 | -0.1594 | No | ||
| 62 | TJP2 | 7173 | 0.685 | -0.1593 | No | ||
| 63 | CRADD | 7376 | 0.650 | -0.1688 | No | ||
| 64 | PLCB4 | 7397 | 0.646 | -0.1684 | No | ||
| 65 | EHBP1 | 7417 | 0.641 | -0.1680 | No | ||
| 66 | CDC42BPA | 7458 | 0.634 | -0.1688 | No | ||
| 67 | NFKB1 | 7571 | 0.612 | -0.1735 | No | ||
| 68 | GLRB | 7757 | 0.574 | -0.1823 | No | ||
| 69 | TCF7L2 | 7952 | 0.538 | -0.1916 | No | ||
| 70 | SEMA3C | 8031 | 0.525 | -0.1947 | No | ||
| 71 | EPS15 | 8074 | 0.516 | -0.1958 | No | ||
| 72 | FARS2 | 8491 | 0.436 | -0.2175 | No | ||
| 73 | EPS8 | 8493 | 0.436 | -0.2166 | No | ||
| 74 | CENTG2 | 8569 | 0.422 | -0.2197 | No | ||
| 75 | FGF5 | 8688 | 0.401 | -0.2252 | No | ||
| 76 | PTPN2 | 8723 | 0.395 | -0.2262 | No | ||
| 77 | GBE1 | 8745 | 0.390 | -0.2264 | No | ||
| 78 | PLCE1 | 8773 | 0.386 | -0.2270 | No | ||
| 79 | SMURF2 | 8782 | 0.384 | -0.2266 | No | ||
| 80 | ACSL3 | 8893 | 0.365 | -0.2318 | No | ||
| 81 | KIF5C | 8907 | 0.362 | -0.2316 | No | ||
| 82 | SLC25A12 | 9077 | 0.327 | -0.2401 | No | ||
| 83 | RRAS2 | 9117 | 0.319 | -0.2415 | No | ||
| 84 | TUBGCP3 | 9120 | 0.318 | -0.2409 | No | ||
| 85 | MSH2 | 9177 | 0.304 | -0.2433 | No | ||
| 86 | TIPARP | 9234 | 0.291 | -0.2457 | No | ||
| 87 | ADAM10 | 9326 | 0.275 | -0.2500 | No | ||
| 88 | PLEKHC1 | 9336 | 0.273 | -0.2499 | No | ||
| 89 | ITSN1 | 9422 | 0.250 | -0.2539 | No | ||
| 90 | SEPT9 | 9569 | 0.218 | -0.2614 | No | ||
| 91 | SCHIP1 | 9714 | 0.185 | -0.2688 | No | ||
| 92 | STK39 | 9789 | 0.168 | -0.2725 | No | ||
| 93 | PRKCA | 9829 | 0.161 | -0.2743 | No | ||
| 94 | NRIP1 | 9881 | 0.150 | -0.2767 | No | ||
| 95 | SLIT2 | 9923 | 0.143 | -0.2786 | No | ||
| 96 | THBS2 | 9967 | 0.134 | -0.2806 | No | ||
| 97 | KLF6 | 9982 | 0.131 | -0.2811 | No | ||
| 98 | STAU2 | 10064 | 0.116 | -0.2853 | No | ||
| 99 | STK24 | 10285 | 0.069 | -0.2971 | No | ||
| 100 | ADARB1 | 10304 | 0.063 | -0.2980 | No | ||
| 101 | NMT2 | 10660 | -0.017 | -0.3173 | No | ||
| 102 | DPYD | 10756 | -0.041 | -0.3224 | No | ||
| 103 | FBXW11 | 10879 | -0.066 | -0.3289 | No | ||
| 104 | XPO1 | 10884 | -0.068 | -0.3289 | No | ||
| 105 | MTX2 | 11121 | -0.119 | -0.3416 | No | ||
| 106 | CENPF | 11128 | -0.120 | -0.3416 | No | ||
| 107 | USP13 | 11347 | -0.171 | -0.3531 | No | ||
| 108 | TRIM37 | 11348 | -0.171 | -0.3527 | No | ||
| 109 | CLASP2 | 11399 | -0.184 | -0.3550 | No | ||
| 110 | VEGFC | 11480 | -0.206 | -0.3589 | No | ||
| 111 | DDX10 | 11510 | -0.211 | -0.3600 | No | ||
| 112 | EPHA4 | 11818 | -0.279 | -0.3761 | No | ||
| 113 | PIK3C2A | 11952 | -0.312 | -0.3827 | No | ||
| 114 | PKD2 | 12240 | -0.380 | -0.3975 | No | ||
| 115 | NPEPPS | 12259 | -0.384 | -0.3976 | No | ||
| 116 | KPNA3 | 12330 | -0.403 | -0.4005 | No | ||
| 117 | TLE4 | 12360 | -0.409 | -0.4011 | No | ||
| 118 | NCOA3 | 12515 | -0.452 | -0.4085 | No | ||
| 119 | RAB6IP1 | 12618 | -0.477 | -0.4130 | No | ||
| 120 | PHF21A | 12693 | -0.494 | -0.4159 | No | ||
| 121 | GOLGA4 | 12713 | -0.499 | -0.4158 | No | ||
| 122 | IFRD1 | 12779 | -0.515 | -0.4182 | No | ||
| 123 | THRAP2 | 12787 | -0.516 | -0.4174 | No | ||
| 124 | PRIM2A | 12842 | -0.531 | -0.4191 | No | ||
| 125 | ZFYVE9 | 12847 | -0.532 | -0.4181 | No | ||
| 126 | F3 | 13007 | -0.576 | -0.4255 | No | ||
| 127 | KIAA0427 | 13036 | -0.583 | -0.4257 | No | ||
| 128 | AUH | 13090 | -0.600 | -0.4272 | No | ||
| 129 | CLOCK | 13167 | -0.620 | -0.4299 | No | ||
| 130 | RGS20 | 13203 | -0.629 | -0.4304 | No | ||
| 131 | DYRK1A | 13236 | -0.637 | -0.4307 | No | ||
| 132 | ORC2L | 13315 | -0.665 | -0.4335 | No | ||
| 133 | ADCY9 | 13322 | -0.667 | -0.4323 | No | ||
| 134 | NEK7 | 13361 | -0.680 | -0.4328 | No | ||
| 135 | IL7R | 13373 | -0.683 | -0.4318 | No | ||
| 136 | LRPPRC | 13390 | -0.687 | -0.4311 | No | ||
| 137 | USP9X | 13424 | -0.697 | -0.4314 | No | ||
| 138 | RPGR | 13454 | -0.706 | -0.4313 | No | ||
| 139 | CLASP1 | 13461 | -0.707 | -0.4300 | No | ||
| 140 | ZNF148 | 13471 | -0.708 | -0.4289 | No | ||
| 141 | MYO9B | 13501 | -0.718 | -0.4289 | No | ||
| 142 | PHLPP | 13546 | -0.732 | -0.4296 | No | ||
| 143 | R3HDM1 | 13572 | -0.740 | -0.4293 | No | ||
| 144 | STARD13 | 13801 | -0.811 | -0.4399 | No | ||
| 145 | DNMBP | 13844 | -0.821 | -0.4403 | No | ||
| 146 | SATB2 | 13926 | -0.842 | -0.4428 | No | ||
| 147 | NFYC | 13982 | -0.858 | -0.4438 | No | ||
| 148 | BCAR3 | 13985 | -0.860 | -0.4420 | No | ||
| 149 | HIVEP1 | 14034 | -0.878 | -0.4426 | No | ||
| 150 | MARK3 | 14066 | -0.889 | -0.4423 | No | ||
| 151 | ATP2C1 | 14098 | -0.902 | -0.4419 | No | ||
| 152 | MYC | 14099 | -0.902 | -0.4399 | No | ||
| 153 | RAD23B | 14126 | -0.911 | -0.4392 | No | ||
| 154 | E2F5 | 14208 | -0.936 | -0.4415 | No | ||
| 155 | XPO6 | 14218 | -0.939 | -0.4398 | No | ||
| 156 | IRAK1BP1 | 14555 | -1.041 | -0.4558 | No | ||
| 157 | PTEN | 14609 | -1.063 | -0.4563 | No | ||
| 158 | LYN | 14620 | -1.067 | -0.4544 | No | ||
| 159 | TBL1X | 14710 | -1.102 | -0.4567 | No | ||
| 160 | DLG1 | 14724 | -1.106 | -0.4549 | No | ||
| 161 | PCNT | 14871 | -1.159 | -0.4602 | No | ||
| 162 | SEC14L1 | 14982 | -1.202 | -0.4635 | Yes | ||
| 163 | PTPRK | 14983 | -1.203 | -0.4608 | Yes | ||
| 164 | GTF2F2 | 15008 | -1.211 | -0.4593 | Yes | ||
| 165 | TOPBP1 | 15048 | -1.224 | -0.4587 | Yes | ||
| 166 | TDRD7 | 15068 | -1.233 | -0.4569 | Yes | ||
| 167 | LASS6 | 15069 | -1.235 | -0.4541 | Yes | ||
| 168 | PARD3 | 15072 | -1.235 | -0.4514 | Yes | ||
| 169 | CASK | 15148 | -1.265 | -0.4526 | Yes | ||
| 170 | LARS2 | 15270 | -1.315 | -0.4562 | Yes | ||
| 171 | SOCS5 | 15286 | -1.320 | -0.4540 | Yes | ||
| 172 | CREBBP | 15329 | -1.334 | -0.4532 | Yes | ||
| 173 | GSK3B | 15391 | -1.361 | -0.4535 | Yes | ||
| 174 | MAN2A1 | 15406 | -1.367 | -0.4511 | Yes | ||
| 175 | MDN1 | 15474 | -1.395 | -0.4516 | Yes | ||
| 176 | CDS2 | 15478 | -1.396 | -0.4486 | Yes | ||
| 177 | PBX3 | 15509 | -1.408 | -0.4470 | Yes | ||
| 178 | SLC25A13 | 15614 | -1.451 | -0.4494 | Yes | ||
| 179 | KLF7 | 15710 | -1.494 | -0.4512 | Yes | ||
| 180 | PICALM | 15769 | -1.519 | -0.4509 | Yes | ||
| 181 | ARFGEF1 | 15799 | -1.532 | -0.4490 | Yes | ||
| 182 | PPM1B | 15820 | -1.541 | -0.4465 | Yes | ||
| 183 | RPS6KA3 | 15856 | -1.562 | -0.4449 | Yes | ||
| 184 | USP12 | 15860 | -1.564 | -0.4415 | Yes | ||
| 185 | PCCA | 15887 | -1.572 | -0.4393 | Yes | ||
| 186 | MTAP | 15900 | -1.579 | -0.4364 | Yes | ||
| 187 | PTPN12 | 15902 | -1.580 | -0.4328 | Yes | ||
| 188 | ZMYND11 | 15939 | -1.594 | -0.4312 | Yes | ||
| 189 | SAMD4A | 16042 | -1.646 | -0.4330 | Yes | ||
| 190 | RBM16 | 16043 | -1.646 | -0.4292 | Yes | ||
| 191 | PCMT1 | 16057 | -1.654 | -0.4262 | Yes | ||
| 192 | MICAL3 | 16145 | -1.690 | -0.4271 | Yes | ||
| 193 | MID1 | 16173 | -1.701 | -0.4247 | Yes | ||
| 194 | PDE8A | 16199 | -1.714 | -0.4221 | Yes | ||
| 195 | CTBP2 | 16208 | -1.718 | -0.4186 | Yes | ||
| 196 | NT5C2 | 16210 | -1.718 | -0.4148 | Yes | ||
| 197 | XRCC4 | 16226 | -1.726 | -0.4117 | Yes | ||
| 198 | MELK | 16238 | -1.730 | -0.4083 | Yes | ||
| 199 | FNDC3A | 16264 | -1.741 | -0.4057 | Yes | ||
| 200 | ATF2 | 16286 | -1.753 | -0.4029 | Yes | ||
| 201 | ORC5L | 16341 | -1.784 | -0.4018 | Yes | ||
| 202 | NUP133 | 16359 | -1.791 | -0.3986 | Yes | ||
| 203 | PAIP1 | 16376 | -1.799 | -0.3954 | Yes | ||
| 204 | RUNX1 | 16383 | -1.803 | -0.3916 | Yes | ||
| 205 | STK3 | 16407 | -1.816 | -0.3887 | Yes | ||
| 206 | PEX3 | 16427 | -1.822 | -0.3856 | Yes | ||
| 207 | SLC4A7 | 16499 | -1.856 | -0.3852 | Yes | ||
| 208 | BARD1 | 16590 | -1.904 | -0.3858 | Yes | ||
| 209 | TRIP12 | 16595 | -1.907 | -0.3817 | Yes | ||
| 210 | MYST4 | 16623 | -1.922 | -0.3788 | Yes | ||
| 211 | TOP2A | 16650 | -1.933 | -0.3758 | Yes | ||
| 212 | UBE2D2 | 16660 | -1.936 | -0.3719 | Yes | ||
| 213 | PPP2R5E | 16688 | -1.953 | -0.3689 | Yes | ||
| 214 | DNAJC13 | 16694 | -1.955 | -0.3647 | Yes | ||
| 215 | TCF12 | 16722 | -1.970 | -0.3617 | Yes | ||
| 216 | MYO10 | 16725 | -1.977 | -0.3573 | Yes | ||
| 217 | ARIH1 | 16744 | -1.986 | -0.3538 | Yes | ||
| 218 | RAD51C | 16817 | -2.028 | -0.3531 | Yes | ||
| 219 | UBL3 | 16825 | -2.031 | -0.3488 | Yes | ||
| 220 | CUL1 | 16836 | -2.035 | -0.3447 | Yes | ||
| 221 | RAB6A | 16886 | -2.059 | -0.3427 | Yes | ||
| 222 | CUTL1 | 16954 | -2.094 | -0.3416 | Yes | ||
| 223 | PPP1R12A | 17014 | -2.128 | -0.3400 | Yes | ||
| 224 | HNRPDL | 17024 | -2.135 | -0.3356 | Yes | ||
| 225 | MAP2K4 | 17047 | -2.144 | -0.3319 | Yes | ||
| 226 | RANBP2 | 17048 | -2.144 | -0.3270 | Yes | ||
| 227 | PAFAH1B1 | 17084 | -2.165 | -0.3240 | Yes | ||
| 228 | PUM2 | 17116 | -2.184 | -0.3207 | Yes | ||
| 229 | SON | 17130 | -2.195 | -0.3164 | Yes | ||
| 230 | DAPK1 | 17149 | -2.205 | -0.3124 | Yes | ||
| 231 | PTK2 | 17189 | -2.224 | -0.3094 | Yes | ||
| 232 | IBTK | 17197 | -2.229 | -0.3047 | Yes | ||
| 233 | PJA2 | 17221 | -2.243 | -0.3009 | Yes | ||
| 234 | NBN | 17226 | -2.246 | -0.2960 | Yes | ||
| 235 | ATRN | 17234 | -2.250 | -0.2912 | Yes | ||
| 236 | RB1 | 17323 | -2.301 | -0.2908 | Yes | ||
| 237 | OPA1 | 17382 | -2.339 | -0.2886 | Yes | ||
| 238 | ZZZ3 | 17512 | -2.418 | -0.2902 | Yes | ||
| 239 | SEC24B | 17516 | -2.420 | -0.2848 | Yes | ||
| 240 | FUT8 | 17568 | -2.449 | -0.2820 | Yes | ||
| 241 | TRIM33 | 17582 | -2.459 | -0.2771 | Yes | ||
| 242 | MAP2K1 | 17659 | -2.510 | -0.2755 | Yes | ||
| 243 | MALT1 | 17690 | -2.531 | -0.2714 | Yes | ||
| 244 | TLK1 | 17754 | -2.574 | -0.2690 | Yes | ||
| 245 | RNGTT | 17757 | -2.575 | -0.2632 | Yes | ||
| 246 | STRN3 | 17765 | -2.578 | -0.2577 | Yes | ||
| 247 | WDR7 | 17774 | -2.584 | -0.2523 | Yes | ||
| 248 | ATR | 17777 | -2.586 | -0.2465 | Yes | ||
| 249 | TLK2 | 17816 | -2.616 | -0.2426 | Yes | ||
| 250 | MBNL1 | 17818 | -2.617 | -0.2367 | Yes | ||
| 251 | CDH2 | 17829 | -2.624 | -0.2313 | Yes | ||
| 252 | PKN2 | 17845 | -2.641 | -0.2261 | Yes | ||
| 253 | EIF4G3 | 17856 | -2.652 | -0.2206 | Yes | ||
| 254 | FCHSD2 | 17866 | -2.659 | -0.2150 | Yes | ||
| 255 | SFRS3 | 17871 | -2.665 | -0.2092 | Yes | ||
| 256 | NUMB | 17891 | -2.678 | -0.2041 | Yes | ||
| 257 | CNOT2 | 17907 | -2.690 | -0.1988 | Yes | ||
| 258 | SOS2 | 17945 | -2.716 | -0.1946 | Yes | ||
| 259 | OGT | 17951 | -2.718 | -0.1887 | Yes | ||
| 260 | NVL | 17967 | -2.737 | -0.1833 | Yes | ||
| 261 | ANKMY2 | 17993 | -2.757 | -0.1784 | Yes | ||
| 262 | CDK8 | 17994 | -2.758 | -0.1721 | Yes | ||
| 263 | STAG1 | 17999 | -2.762 | -0.1660 | Yes | ||
| 264 | PARG | 18020 | -2.785 | -0.1607 | Yes | ||
| 265 | PRKACB | 18025 | -2.788 | -0.1546 | Yes | ||
| 266 | ITSN2 | 18042 | -2.802 | -0.1491 | Yes | ||
| 267 | ATXN2 | 18057 | -2.820 | -0.1434 | Yes | ||
| 268 | DHX15 | 18058 | -2.822 | -0.1370 | Yes | ||
| 269 | PRMT3 | 18060 | -2.823 | -0.1306 | Yes | ||
| 270 | ZNF638 | 18075 | -2.833 | -0.1249 | Yes | ||
| 271 | EIF4E | 18085 | -2.842 | -0.1189 | Yes | ||
| 272 | GMPS | 18106 | -2.868 | -0.1135 | Yes | ||
| 273 | RASA1 | 18140 | -2.908 | -0.1087 | Yes | ||
| 274 | MKLN1 | 18143 | -2.908 | -0.1022 | Yes | ||
| 275 | PIK3C3 | 18161 | -2.927 | -0.0964 | Yes | ||
| 276 | UBE2G1 | 18166 | -2.934 | -0.0899 | Yes | ||
| 277 | UPF2 | 18242 | -3.039 | -0.0871 | Yes | ||
| 278 | GRK5 | 18273 | -3.087 | -0.0817 | Yes | ||
| 279 | SRPK2 | 18286 | -3.115 | -0.0753 | Yes | ||
| 280 | REV3L | 18312 | -3.147 | -0.0695 | Yes | ||
| 281 | MPHOSPH1 | 18370 | -3.248 | -0.0652 | Yes | ||
| 282 | ROCK2 | 18399 | -3.300 | -0.0592 | Yes | ||
| 283 | ANKRD17 | 18423 | -3.382 | -0.0527 | Yes | ||
| 284 | CHD1 | 18463 | -3.482 | -0.0469 | Yes | ||
| 285 | ATP11B | 18483 | -3.537 | -0.0399 | Yes | ||
| 286 | SSBP2 | 18492 | -3.556 | -0.0322 | Yes | ||
| 287 | APPBP2 | 18510 | -3.674 | -0.0248 | Yes | ||
| 288 | CDYL | 18552 | -3.934 | -0.0181 | Yes | ||
| 289 | SMC5 | 18564 | -4.046 | -0.0095 | Yes | ||
| 290 | IL1RAP | 18610 | -5.370 | 0.0003 | Yes |
