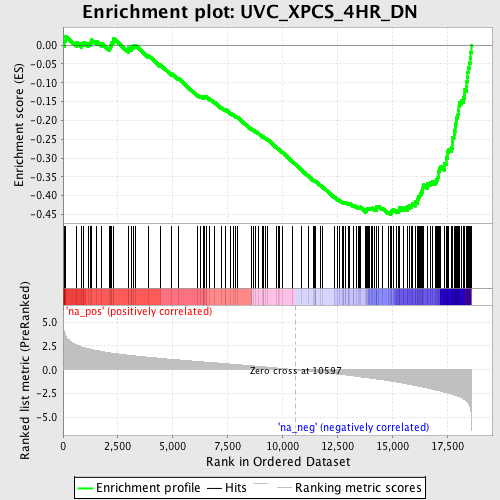
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | UVC_XPCS_4HR_DN |
| Enrichment Score (ES) | -0.4500801 |
| Normalized Enrichment Score (NES) | -2.2388418 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.0055122E-4 |
| FWER p-Value | 0.0070 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GNAQ | 64 | 3.955 | 0.0122 | No | ||
| 2 | DLC1 | 109 | 3.570 | 0.0240 | No | ||
| 3 | LHFPL2 | 613 | 2.599 | 0.0070 | No | ||
| 4 | CREB1 | 854 | 2.389 | 0.0035 | No | ||
| 5 | TRIM2 | 943 | 2.323 | 0.0079 | No | ||
| 6 | EGFR | 1159 | 2.187 | 0.0049 | No | ||
| 7 | DOCK4 | 1257 | 2.133 | 0.0081 | No | ||
| 8 | ZHX3 | 1305 | 2.107 | 0.0139 | No | ||
| 9 | FYN | 1528 | 2.012 | 0.0098 | No | ||
| 10 | RNF13 | 1757 | 1.920 | 0.0051 | No | ||
| 11 | EXT1 | 2128 | 1.776 | -0.0079 | No | ||
| 12 | IGF1R | 2150 | 1.768 | -0.0021 | No | ||
| 13 | BICD1 | 2188 | 1.754 | 0.0029 | No | ||
| 14 | PRR3 | 2226 | 1.741 | 0.0078 | No | ||
| 15 | CBLB | 2285 | 1.721 | 0.0114 | No | ||
| 16 | FGF2 | 2313 | 1.711 | 0.0168 | No | ||
| 17 | BTRC | 2982 | 1.526 | -0.0134 | No | ||
| 18 | MTMR3 | 2984 | 1.526 | -0.0074 | No | ||
| 19 | CENPE | 3121 | 1.491 | -0.0089 | No | ||
| 20 | TLE1 | 3135 | 1.487 | -0.0037 | No | ||
| 21 | SRGAP2 | 3213 | 1.466 | -0.0020 | No | ||
| 22 | ANKS1A | 3286 | 1.445 | -0.0002 | No | ||
| 23 | MYST3 | 3903 | 1.304 | -0.0284 | No | ||
| 24 | ARHGEF7 | 4422 | 1.194 | -0.0518 | No | ||
| 25 | TIAM1 | 4950 | 1.089 | -0.0760 | No | ||
| 26 | BHLHB2 | 5252 | 1.032 | -0.0883 | No | ||
| 27 | RGS7 | 6140 | 0.866 | -0.1329 | No | ||
| 28 | CD2AP | 6276 | 0.840 | -0.1369 | No | ||
| 29 | TGFB2 | 6387 | 0.822 | -0.1396 | No | ||
| 30 | ZCCHC11 | 6410 | 0.817 | -0.1376 | No | ||
| 31 | HOXB2 | 6450 | 0.810 | -0.1365 | No | ||
| 32 | NCK1 | 6517 | 0.798 | -0.1369 | No | ||
| 33 | TSPAN5 | 6687 | 0.766 | -0.1430 | No | ||
| 34 | HIRA | 6908 | 0.727 | -0.1520 | No | ||
| 35 | WNT5A | 7213 | 0.677 | -0.1658 | No | ||
| 36 | ACVR1 | 7384 | 0.648 | -0.1725 | No | ||
| 37 | EHBP1 | 7417 | 0.641 | -0.1717 | No | ||
| 38 | CTGF | 7634 | 0.599 | -0.1810 | No | ||
| 39 | GLRB | 7757 | 0.574 | -0.1854 | No | ||
| 40 | ARHGAP12 | 7867 | 0.555 | -0.1891 | No | ||
| 41 | TCF7L2 | 7952 | 0.538 | -0.1915 | No | ||
| 42 | CENTG2 | 8569 | 0.422 | -0.2232 | No | ||
| 43 | NFIB | 8575 | 0.420 | -0.2218 | No | ||
| 44 | FGF5 | 8688 | 0.401 | -0.2263 | No | ||
| 45 | SMURF2 | 8782 | 0.384 | -0.2298 | No | ||
| 46 | ACSL3 | 8893 | 0.365 | -0.2343 | No | ||
| 47 | SLC25A12 | 9077 | 0.327 | -0.2429 | No | ||
| 48 | RRAS2 | 9117 | 0.319 | -0.2438 | No | ||
| 49 | TUBGCP3 | 9120 | 0.318 | -0.2426 | No | ||
| 50 | TIPARP | 9234 | 0.291 | -0.2476 | No | ||
| 51 | PLEKHC1 | 9336 | 0.273 | -0.2520 | No | ||
| 52 | SCHIP1 | 9714 | 0.185 | -0.2717 | No | ||
| 53 | BMPR1A | 9798 | 0.166 | -0.2756 | No | ||
| 54 | NRIP1 | 9881 | 0.150 | -0.2794 | No | ||
| 55 | KLF6 | 9982 | 0.131 | -0.2843 | No | ||
| 56 | TERF1 | 10434 | 0.034 | -0.3086 | No | ||
| 57 | FBXW11 | 10879 | -0.066 | -0.3324 | No | ||
| 58 | SPEN | 11161 | -0.131 | -0.3471 | No | ||
| 59 | CLASP2 | 11399 | -0.184 | -0.3592 | No | ||
| 60 | FBXL7 | 11441 | -0.197 | -0.3607 | No | ||
| 61 | VEGFC | 11480 | -0.206 | -0.3619 | No | ||
| 62 | DDX10 | 11510 | -0.211 | -0.3627 | No | ||
| 63 | TAF4 | 11511 | -0.211 | -0.3618 | No | ||
| 64 | THRAP1 | 11739 | -0.262 | -0.3731 | No | ||
| 65 | EPHA4 | 11818 | -0.279 | -0.3762 | No | ||
| 66 | TLE4 | 12360 | -0.409 | -0.4039 | No | ||
| 67 | NCOA3 | 12515 | -0.452 | -0.4105 | No | ||
| 68 | CENPC1 | 12604 | -0.474 | -0.4134 | No | ||
| 69 | GNAI1 | 12720 | -0.500 | -0.4176 | No | ||
| 70 | IFRD1 | 12779 | -0.515 | -0.4187 | No | ||
| 71 | THRAP2 | 12787 | -0.516 | -0.4171 | No | ||
| 72 | ZFYVE9 | 12847 | -0.532 | -0.4182 | No | ||
| 73 | ZHX2 | 12884 | -0.541 | -0.4180 | No | ||
| 74 | RCOR1 | 13002 | -0.575 | -0.4220 | No | ||
| 75 | F3 | 13007 | -0.576 | -0.4200 | No | ||
| 76 | TGFBR3 | 13065 | -0.593 | -0.4207 | No | ||
| 77 | DYRK1A | 13236 | -0.637 | -0.4274 | No | ||
| 78 | MTF2 | 13249 | -0.642 | -0.4255 | No | ||
| 79 | CENTD1 | 13377 | -0.685 | -0.4297 | No | ||
| 80 | ZNF148 | 13471 | -0.708 | -0.4319 | No | ||
| 81 | KLF10 | 13524 | -0.725 | -0.4318 | No | ||
| 82 | PHLPP | 13546 | -0.732 | -0.4301 | No | ||
| 83 | STARD13 | 13801 | -0.811 | -0.4406 | No | ||
| 84 | PCTK2 | 13824 | -0.817 | -0.4386 | No | ||
| 85 | DNMBP | 13844 | -0.821 | -0.4364 | No | ||
| 86 | CENPA | 13875 | -0.829 | -0.4347 | No | ||
| 87 | SATB2 | 13926 | -0.842 | -0.4341 | No | ||
| 88 | BCAR3 | 13985 | -0.860 | -0.4338 | No | ||
| 89 | MARK3 | 14066 | -0.889 | -0.4346 | No | ||
| 90 | MYC | 14099 | -0.902 | -0.4328 | No | ||
| 91 | E2F5 | 14208 | -0.936 | -0.4349 | No | ||
| 92 | PIP5K3 | 14280 | -0.958 | -0.4350 | No | ||
| 93 | SHB | 14297 | -0.963 | -0.4320 | No | ||
| 94 | NUP153 | 14299 | -0.964 | -0.4283 | No | ||
| 95 | EIF2C2 | 14366 | -0.986 | -0.4279 | No | ||
| 96 | IRAK1BP1 | 14555 | -1.041 | -0.4340 | No | ||
| 97 | SOS1 | 14822 | -1.141 | -0.4439 | No | ||
| 98 | PVRL3 | 14937 | -1.182 | -0.4454 | Yes | ||
| 99 | IL6 | 14967 | -1.195 | -0.4422 | Yes | ||
| 100 | PTPRK | 14983 | -1.203 | -0.4383 | Yes | ||
| 101 | TOPBP1 | 15048 | -1.224 | -0.4369 | Yes | ||
| 102 | USP15 | 15201 | -1.286 | -0.4400 | Yes | ||
| 103 | SOCS5 | 15286 | -1.320 | -0.4394 | Yes | ||
| 104 | SWAP70 | 15316 | -1.331 | -0.4357 | Yes | ||
| 105 | CREBBP | 15329 | -1.334 | -0.4310 | Yes | ||
| 106 | PBX3 | 15509 | -1.408 | -0.4352 | Yes | ||
| 107 | KIF2A | 15533 | -1.417 | -0.4308 | Yes | ||
| 108 | UGCG | 15685 | -1.484 | -0.4331 | Yes | ||
| 109 | KLF7 | 15710 | -1.494 | -0.4285 | Yes | ||
| 110 | ARFGEF1 | 15799 | -1.532 | -0.4272 | Yes | ||
| 111 | MTAP | 15900 | -1.579 | -0.4263 | Yes | ||
| 112 | PTPN12 | 15902 | -1.580 | -0.4201 | Yes | ||
| 113 | SAMD4A | 16042 | -1.646 | -0.4211 | Yes | ||
| 114 | RBM16 | 16043 | -1.646 | -0.4146 | Yes | ||
| 115 | MICAL3 | 16145 | -1.690 | -0.4134 | Yes | ||
| 116 | MID1 | 16173 | -1.701 | -0.4081 | Yes | ||
| 117 | CTBP2 | 16208 | -1.718 | -0.4032 | Yes | ||
| 118 | FNDC3A | 16264 | -1.741 | -0.3992 | Yes | ||
| 119 | ATF2 | 16286 | -1.753 | -0.3934 | Yes | ||
| 120 | ORC5L | 16341 | -1.784 | -0.3893 | Yes | ||
| 121 | RUNX1 | 16383 | -1.803 | -0.3844 | Yes | ||
| 122 | AKAP10 | 16400 | -1.812 | -0.3781 | Yes | ||
| 123 | STK3 | 16407 | -1.816 | -0.3712 | Yes | ||
| 124 | BARD1 | 16590 | -1.904 | -0.3735 | Yes | ||
| 125 | MYST4 | 16623 | -1.922 | -0.3676 | Yes | ||
| 126 | MYO10 | 16725 | -1.977 | -0.3653 | Yes | ||
| 127 | NCOA1 | 16840 | -2.036 | -0.3634 | Yes | ||
| 128 | CUTL1 | 16954 | -2.094 | -0.3612 | Yes | ||
| 129 | PPP1R12A | 17014 | -2.128 | -0.3560 | Yes | ||
| 130 | PAFAH1B1 | 17084 | -2.165 | -0.3511 | Yes | ||
| 131 | APRIN | 17114 | -2.181 | -0.3441 | Yes | ||
| 132 | PUM2 | 17116 | -2.184 | -0.3355 | Yes | ||
| 133 | DAPK1 | 17149 | -2.205 | -0.3285 | Yes | ||
| 134 | PTK2 | 17189 | -2.224 | -0.3218 | Yes | ||
| 135 | CYR61 | 17381 | -2.338 | -0.3229 | Yes | ||
| 136 | ZNF292 | 17383 | -2.339 | -0.3136 | Yes | ||
| 137 | RBM39 | 17472 | -2.390 | -0.3089 | Yes | ||
| 138 | CNOT4 | 17473 | -2.390 | -0.2995 | Yes | ||
| 139 | ZZZ3 | 17512 | -2.418 | -0.2919 | Yes | ||
| 140 | SEC24B | 17516 | -2.420 | -0.2825 | Yes | ||
| 141 | TRIM33 | 17582 | -2.459 | -0.2763 | Yes | ||
| 142 | MALT1 | 17690 | -2.531 | -0.2721 | Yes | ||
| 143 | TLK1 | 17754 | -2.574 | -0.2653 | Yes | ||
| 144 | RNGTT | 17757 | -2.575 | -0.2552 | Yes | ||
| 145 | STRN3 | 17765 | -2.578 | -0.2453 | Yes | ||
| 146 | TLK2 | 17816 | -2.616 | -0.2377 | Yes | ||
| 147 | BLM | 17825 | -2.619 | -0.2277 | Yes | ||
| 148 | FCHSD2 | 17866 | -2.659 | -0.2194 | Yes | ||
| 149 | MARCH7 | 17877 | -2.667 | -0.2094 | Yes | ||
| 150 | CNOT2 | 17907 | -2.690 | -0.2003 | Yes | ||
| 151 | SOS2 | 17945 | -2.716 | -0.1915 | Yes | ||
| 152 | CDK8 | 17994 | -2.758 | -0.1832 | Yes | ||
| 153 | PARG | 18020 | -2.785 | -0.1735 | Yes | ||
| 154 | ITSN2 | 18042 | -2.802 | -0.1635 | Yes | ||
| 155 | ATXN2 | 18057 | -2.820 | -0.1531 | Yes | ||
| 156 | RASA1 | 18140 | -2.908 | -0.1460 | Yes | ||
| 157 | UPF2 | 18242 | -3.039 | -0.1395 | Yes | ||
| 158 | SRPK2 | 18286 | -3.115 | -0.1295 | Yes | ||
| 159 | REV3L | 18312 | -3.147 | -0.1184 | Yes | ||
| 160 | MECP2 | 18396 | -3.287 | -0.1098 | Yes | ||
| 161 | WDR37 | 18402 | -3.309 | -0.0970 | Yes | ||
| 162 | ANKRD17 | 18423 | -3.382 | -0.0847 | Yes | ||
| 163 | AKAP9 | 18438 | -3.420 | -0.0719 | Yes | ||
| 164 | CHD1 | 18463 | -3.482 | -0.0594 | Yes | ||
| 165 | APPBP2 | 18510 | -3.674 | -0.0473 | Yes | ||
| 166 | CDYL | 18552 | -3.934 | -0.0340 | Yes | ||
| 167 | STAM | 18565 | -4.049 | -0.0186 | Yes | ||
| 168 | IL1RAP | 18610 | -5.370 | 0.0003 | Yes |
