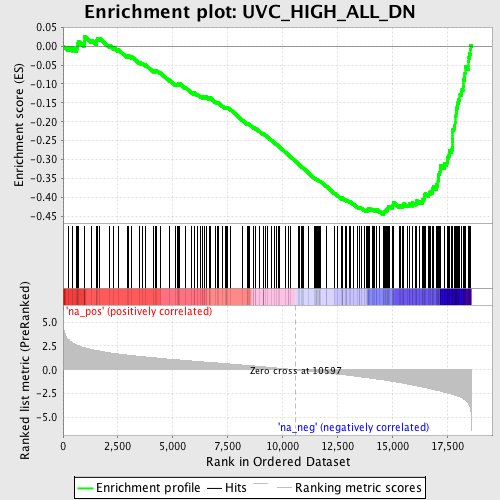
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | UVC_HIGH_ALL_DN |
| Enrichment Score (ES) | -0.44542426 |
| Normalized Enrichment Score (NES) | -2.3257036 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.383894E-5 |
| FWER p-Value | 0.0010 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DUSP1 | 234 | 3.185 | -0.0028 | No | ||
| 2 | UNG | 422 | 2.820 | -0.0042 | No | ||
| 3 | LHFPL2 | 613 | 2.599 | -0.0064 | No | ||
| 4 | PHC2 | 635 | 2.579 | 0.0005 | No | ||
| 5 | HAS2 | 673 | 2.542 | 0.0064 | No | ||
| 6 | ARF6 | 700 | 2.511 | 0.0128 | No | ||
| 7 | FZD2 | 957 | 2.314 | 0.0061 | No | ||
| 8 | IGF2BP3 | 965 | 2.304 | 0.0129 | No | ||
| 9 | PTPN1 | 975 | 2.295 | 0.0195 | No | ||
| 10 | TWIST1 | 977 | 2.293 | 0.0266 | No | ||
| 11 | ZHX3 | 1305 | 2.107 | 0.0154 | No | ||
| 12 | HDHD1A | 1508 | 2.025 | 0.0107 | No | ||
| 13 | FYN | 1528 | 2.012 | 0.0160 | No | ||
| 14 | SPRED2 | 1545 | 2.005 | 0.0213 | No | ||
| 15 | FADD | 1663 | 1.958 | 0.0211 | No | ||
| 16 | CEBPD | 2113 | 1.784 | 0.0022 | No | ||
| 17 | SMAD3 | 2315 | 1.711 | -0.0034 | No | ||
| 18 | PKP4 | 2513 | 1.659 | -0.0089 | No | ||
| 19 | ACVR1B | 2923 | 1.541 | -0.0264 | No | ||
| 20 | MTMR3 | 2984 | 1.526 | -0.0249 | No | ||
| 21 | TLE1 | 3135 | 1.487 | -0.0284 | No | ||
| 22 | NR2F2 | 3482 | 1.401 | -0.0428 | No | ||
| 23 | ZFP36L2 | 3606 | 1.375 | -0.0452 | No | ||
| 24 | PMP22 | 3751 | 1.339 | -0.0489 | No | ||
| 25 | HERPUD1 | 4113 | 1.265 | -0.0646 | No | ||
| 26 | FZD7 | 4190 | 1.249 | -0.0648 | No | ||
| 27 | CTDSP2 | 4233 | 1.239 | -0.0633 | No | ||
| 28 | ARHGEF7 | 4422 | 1.194 | -0.0698 | No | ||
| 29 | AGTR1 | 4846 | 1.108 | -0.0893 | No | ||
| 30 | FOSL2 | 5125 | 1.056 | -0.1011 | No | ||
| 31 | IL4R | 5194 | 1.042 | -0.1016 | No | ||
| 32 | BHLHB2 | 5252 | 1.032 | -0.1015 | No | ||
| 33 | TRIP10 | 5255 | 1.032 | -0.0984 | No | ||
| 34 | TRIB2 | 5321 | 1.019 | -0.0987 | No | ||
| 35 | CXCL12 | 5563 | 0.972 | -0.1088 | No | ||
| 36 | ARID5B | 5848 | 0.920 | -0.1214 | No | ||
| 37 | MLX | 5968 | 0.898 | -0.1250 | No | ||
| 38 | IFNGR2 | 5978 | 0.896 | -0.1227 | No | ||
| 39 | PARP1 | 6146 | 0.865 | -0.1291 | No | ||
| 40 | CD2AP | 6276 | 0.840 | -0.1335 | No | ||
| 41 | PPP2R5C | 6354 | 0.829 | -0.1351 | No | ||
| 42 | CITED2 | 6359 | 0.828 | -0.1328 | No | ||
| 43 | HOXB2 | 6450 | 0.810 | -0.1351 | No | ||
| 44 | CDK7 | 6460 | 0.809 | -0.1331 | No | ||
| 45 | NCK1 | 6517 | 0.798 | -0.1337 | No | ||
| 46 | SRF | 6658 | 0.772 | -0.1389 | No | ||
| 47 | TSPAN5 | 6687 | 0.766 | -0.1380 | No | ||
| 48 | STX3 | 6700 | 0.764 | -0.1363 | No | ||
| 49 | POLE2 | 6949 | 0.721 | -0.1475 | No | ||
| 50 | LSM2 | 7033 | 0.708 | -0.1498 | No | ||
| 51 | RHOB | 7066 | 0.704 | -0.1494 | No | ||
| 52 | BACH1 | 7275 | 0.667 | -0.1586 | No | ||
| 53 | SIAH2 | 7405 | 0.645 | -0.1636 | No | ||
| 54 | DLX2 | 7443 | 0.638 | -0.1636 | No | ||
| 55 | ADM | 7447 | 0.636 | -0.1618 | No | ||
| 56 | STXBP3 | 7481 | 0.629 | -0.1617 | No | ||
| 57 | CTGF | 7634 | 0.599 | -0.1680 | No | ||
| 58 | NTF3 | 8188 | 0.495 | -0.1966 | No | ||
| 59 | GATA2 | 8387 | 0.460 | -0.2059 | No | ||
| 60 | GREM1 | 8425 | 0.451 | -0.2065 | No | ||
| 61 | MET | 8434 | 0.448 | -0.2056 | No | ||
| 62 | EPS8 | 8493 | 0.436 | -0.2073 | No | ||
| 63 | RRS1 | 8679 | 0.402 | -0.2162 | No | ||
| 64 | FGF5 | 8688 | 0.401 | -0.2153 | No | ||
| 65 | CKS2 | 8780 | 0.384 | -0.2191 | No | ||
| 66 | SMURF2 | 8782 | 0.384 | -0.2180 | No | ||
| 67 | MFAP1 | 8942 | 0.354 | -0.2255 | No | ||
| 68 | RRAS2 | 9117 | 0.319 | -0.2340 | No | ||
| 69 | TUBGCP3 | 9120 | 0.318 | -0.2331 | No | ||
| 70 | DDIT4 | 9125 | 0.317 | -0.2323 | No | ||
| 71 | NFIL3 | 9133 | 0.315 | -0.2317 | No | ||
| 72 | TIPARP | 9234 | 0.291 | -0.2362 | No | ||
| 73 | PLEKHC1 | 9336 | 0.273 | -0.2409 | No | ||
| 74 | TNFAIP3 | 9502 | 0.232 | -0.2491 | No | ||
| 75 | EDG2 | 9623 | 0.206 | -0.2550 | No | ||
| 76 | SCHIP1 | 9714 | 0.185 | -0.2593 | No | ||
| 77 | BMPR1A | 9798 | 0.166 | -0.2633 | No | ||
| 78 | NRIP1 | 9881 | 0.150 | -0.2673 | No | ||
| 79 | FHL5 | 10156 | 0.096 | -0.2819 | No | ||
| 80 | PDGFRA | 10254 | 0.075 | -0.2869 | No | ||
| 81 | STK24 | 10285 | 0.069 | -0.2884 | No | ||
| 82 | ARL4C | 10378 | 0.048 | -0.2932 | No | ||
| 83 | PNN | 10743 | -0.037 | -0.3129 | No | ||
| 84 | GNE | 10758 | -0.041 | -0.3135 | No | ||
| 85 | XPO1 | 10884 | -0.068 | -0.3201 | No | ||
| 86 | UAP1 | 10887 | -0.069 | -0.3200 | No | ||
| 87 | FBXO46 | 10935 | -0.081 | -0.3223 | No | ||
| 88 | SPEN | 11161 | -0.131 | -0.3341 | No | ||
| 89 | GBAS | 11472 | -0.205 | -0.3504 | No | ||
| 90 | VEGFC | 11480 | -0.206 | -0.3501 | No | ||
| 91 | SMAD7 | 11555 | -0.219 | -0.3534 | No | ||
| 92 | RND3 | 11559 | -0.221 | -0.3529 | No | ||
| 93 | USP10 | 11574 | -0.222 | -0.3530 | No | ||
| 94 | RAF1 | 11635 | -0.236 | -0.3555 | No | ||
| 95 | YY1 | 11680 | -0.245 | -0.3571 | No | ||
| 96 | IRS1 | 11696 | -0.249 | -0.3572 | No | ||
| 97 | STX7 | 11725 | -0.258 | -0.3579 | No | ||
| 98 | CASP3 | 11982 | -0.320 | -0.3708 | No | ||
| 99 | KLF9 | 12007 | -0.325 | -0.3711 | No | ||
| 100 | TLE4 | 12360 | -0.409 | -0.3890 | No | ||
| 101 | NCOA3 | 12515 | -0.452 | -0.3959 | No | ||
| 102 | PDE4B | 12666 | -0.489 | -0.4026 | No | ||
| 103 | GNAI1 | 12720 | -0.500 | -0.4039 | No | ||
| 104 | TARDBP | 12728 | -0.503 | -0.4027 | No | ||
| 105 | DR1 | 12734 | -0.503 | -0.4014 | No | ||
| 106 | FUS | 12866 | -0.537 | -0.4069 | No | ||
| 107 | SSH1 | 12927 | -0.552 | -0.4084 | No | ||
| 108 | E2F3 | 12935 | -0.554 | -0.4070 | No | ||
| 109 | TPST1 | 13039 | -0.585 | -0.4108 | No | ||
| 110 | IDI1 | 13080 | -0.597 | -0.4111 | No | ||
| 111 | DYRK1A | 13236 | -0.637 | -0.4176 | No | ||
| 112 | THBS1 | 13426 | -0.697 | -0.4257 | No | ||
| 113 | DICER1 | 13505 | -0.720 | -0.4277 | No | ||
| 114 | TRIM32 | 13520 | -0.724 | -0.4262 | No | ||
| 115 | COIL | 13604 | -0.753 | -0.4284 | No | ||
| 116 | PODXL | 13747 | -0.798 | -0.4336 | No | ||
| 117 | ITGA6 | 13828 | -0.818 | -0.4354 | No | ||
| 118 | CENPA | 13875 | -0.829 | -0.4353 | No | ||
| 119 | SLC20A2 | 13883 | -0.831 | -0.4331 | No | ||
| 120 | SATB2 | 13926 | -0.842 | -0.4328 | No | ||
| 121 | CDC6 | 13931 | -0.843 | -0.4304 | No | ||
| 122 | BCAR3 | 13985 | -0.860 | -0.4306 | No | ||
| 123 | GEM | 14083 | -0.895 | -0.4331 | No | ||
| 124 | GAS1 | 14136 | -0.913 | -0.4330 | No | ||
| 125 | TNFRSF1A | 14206 | -0.935 | -0.4339 | No | ||
| 126 | PIP5K3 | 14280 | -0.958 | -0.4349 | No | ||
| 127 | NUP153 | 14299 | -0.964 | -0.4328 | No | ||
| 128 | RYBP | 14422 | -1.003 | -0.4363 | No | ||
| 129 | ELF4 | 14590 | -1.055 | -0.4421 | Yes | ||
| 130 | EMG1 | 14614 | -1.065 | -0.4401 | Yes | ||
| 131 | CCNG1 | 14637 | -1.073 | -0.4379 | Yes | ||
| 132 | BLCAP | 14671 | -1.086 | -0.4363 | Yes | ||
| 133 | TBL1X | 14710 | -1.102 | -0.4350 | Yes | ||
| 134 | SERTAD2 | 14742 | -1.113 | -0.4332 | Yes | ||
| 135 | TM4SF1 | 14777 | -1.127 | -0.4315 | Yes | ||
| 136 | WEE1 | 14784 | -1.131 | -0.4283 | Yes | ||
| 137 | CASP8 | 14812 | -1.139 | -0.4262 | Yes | ||
| 138 | STK38 | 14834 | -1.144 | -0.4238 | Yes | ||
| 139 | PFTK1 | 14897 | -1.166 | -0.4236 | Yes | ||
| 140 | RNF4 | 15014 | -1.212 | -0.4261 | Yes | ||
| 141 | PPAP2B | 15021 | -1.214 | -0.4226 | Yes | ||
| 142 | HMGA2 | 15036 | -1.219 | -0.4196 | Yes | ||
| 143 | TOPBP1 | 15048 | -1.224 | -0.4164 | Yes | ||
| 144 | AURKA | 15058 | -1.229 | -0.4130 | Yes | ||
| 145 | SWAP70 | 15316 | -1.331 | -0.4229 | Yes | ||
| 146 | SLBP | 15367 | -1.349 | -0.4214 | Yes | ||
| 147 | ZFAND5 | 15467 | -1.391 | -0.4224 | Yes | ||
| 148 | MAPK14 | 15512 | -1.409 | -0.4204 | Yes | ||
| 149 | KIF2A | 15533 | -1.417 | -0.4171 | Yes | ||
| 150 | UGCG | 15685 | -1.484 | -0.4207 | Yes | ||
| 151 | POGZ | 15766 | -1.518 | -0.4203 | Yes | ||
| 152 | UBTF | 15798 | -1.532 | -0.4172 | Yes | ||
| 153 | HMGB2 | 15908 | -1.582 | -0.4182 | Yes | ||
| 154 | TAF11 | 15919 | -1.587 | -0.4138 | Yes | ||
| 155 | SAMD4A | 16042 | -1.646 | -0.4153 | Yes | ||
| 156 | EMP1 | 16104 | -1.676 | -0.4134 | Yes | ||
| 157 | DDX3X | 16124 | -1.681 | -0.4092 | Yes | ||
| 158 | PHTF2 | 16255 | -1.734 | -0.4109 | Yes | ||
| 159 | RBM5 | 16365 | -1.792 | -0.4112 | Yes | ||
| 160 | RUNX1 | 16383 | -1.803 | -0.4066 | Yes | ||
| 161 | CDR2 | 16445 | -1.829 | -0.4042 | Yes | ||
| 162 | MPHOSPH6 | 16456 | -1.833 | -0.3990 | Yes | ||
| 163 | PRKCD | 16459 | -1.837 | -0.3934 | Yes | ||
| 164 | BMI1 | 16510 | -1.860 | -0.3903 | Yes | ||
| 165 | TOP2A | 16650 | -1.933 | -0.3918 | Yes | ||
| 166 | ATP6V1B2 | 16676 | -1.944 | -0.3872 | Yes | ||
| 167 | MAT2A | 16755 | -1.992 | -0.3852 | Yes | ||
| 168 | NAP1L1 | 16827 | -2.032 | -0.3827 | Yes | ||
| 169 | CUL1 | 16836 | -2.035 | -0.3768 | Yes | ||
| 170 | MAPK6 | 16859 | -2.048 | -0.3716 | Yes | ||
| 171 | SPOP | 17008 | -2.123 | -0.3731 | Yes | ||
| 172 | PPP1R12A | 17014 | -2.128 | -0.3667 | Yes | ||
| 173 | PAFAH1B1 | 17084 | -2.165 | -0.3637 | Yes | ||
| 174 | BRD8 | 17085 | -2.165 | -0.3570 | Yes | ||
| 175 | RAD21 | 17089 | -2.167 | -0.3504 | Yes | ||
| 176 | PUM2 | 17116 | -2.184 | -0.3450 | Yes | ||
| 177 | UBE2N | 17123 | -2.188 | -0.3385 | Yes | ||
| 178 | AMD1 | 17163 | -2.211 | -0.3337 | Yes | ||
| 179 | USP1 | 17179 | -2.219 | -0.3276 | Yes | ||
| 180 | PTK2 | 17189 | -2.224 | -0.3212 | Yes | ||
| 181 | RARS | 17216 | -2.242 | -0.3156 | Yes | ||
| 182 | HMGCR | 17371 | -2.332 | -0.3167 | Yes | ||
| 183 | CYR61 | 17381 | -2.338 | -0.3100 | Yes | ||
| 184 | THUMPD1 | 17496 | -2.408 | -0.3086 | Yes | ||
| 185 | KIF23 | 17511 | -2.417 | -0.3019 | Yes | ||
| 186 | SEC24B | 17516 | -2.420 | -0.2946 | Yes | ||
| 187 | SNX4 | 17553 | -2.441 | -0.2889 | Yes | ||
| 188 | TNFAIP8 | 17597 | -2.475 | -0.2835 | Yes | ||
| 189 | SQLE | 17613 | -2.483 | -0.2766 | Yes | ||
| 190 | NFE2L2 | 17716 | -2.550 | -0.2742 | Yes | ||
| 191 | BNIP2 | 17729 | -2.559 | -0.2669 | Yes | ||
| 192 | NASP | 17744 | -2.569 | -0.2597 | Yes | ||
| 193 | PLK4 | 17746 | -2.570 | -0.2517 | Yes | ||
| 194 | TLK1 | 17754 | -2.574 | -0.2441 | Yes | ||
| 195 | ZBED4 | 17759 | -2.576 | -0.2363 | Yes | ||
| 196 | KPNA2 | 17761 | -2.577 | -0.2283 | Yes | ||
| 197 | STRN3 | 17765 | -2.578 | -0.2204 | Yes | ||
| 198 | TLK2 | 17816 | -2.616 | -0.2150 | Yes | ||
| 199 | CBX3 | 17852 | -2.646 | -0.2087 | Yes | ||
| 200 | KRIT1 | 17861 | -2.657 | -0.2008 | Yes | ||
| 201 | RAB14 | 17870 | -2.665 | -0.1929 | Yes | ||
| 202 | SFRS3 | 17871 | -2.665 | -0.1846 | Yes | ||
| 203 | CNOT2 | 17907 | -2.690 | -0.1782 | Yes | ||
| 204 | GTF2E1 | 17912 | -2.692 | -0.1700 | Yes | ||
| 205 | CRKL | 17947 | -2.716 | -0.1634 | Yes | ||
| 206 | NFYB | 17985 | -2.753 | -0.1568 | Yes | ||
| 207 | CDK8 | 17994 | -2.758 | -0.1487 | Yes | ||
| 208 | MARCKS | 18036 | -2.799 | -0.1422 | Yes | ||
| 209 | EIF4E | 18085 | -2.842 | -0.1359 | Yes | ||
| 210 | CSTF3 | 18086 | -2.844 | -0.1271 | Yes | ||
| 211 | UBE2G1 | 18166 | -2.934 | -0.1222 | Yes | ||
| 212 | RAB1A | 18177 | -2.944 | -0.1136 | Yes | ||
| 213 | CCNA2 | 18230 | -3.023 | -0.1070 | Yes | ||
| 214 | PDCD10 | 18239 | -3.036 | -0.0980 | Yes | ||
| 215 | LBR | 18241 | -3.037 | -0.0886 | Yes | ||
| 216 | DUSP11 | 18288 | -3.117 | -0.0814 | Yes | ||
| 217 | REV3L | 18312 | -3.147 | -0.0728 | Yes | ||
| 218 | PITPNB | 18322 | -3.163 | -0.0635 | Yes | ||
| 219 | RAB11A | 18323 | -3.164 | -0.0536 | Yes | ||
| 220 | CHD1 | 18463 | -3.482 | -0.0503 | Yes | ||
| 221 | ATP2A2 | 18465 | -3.491 | -0.0395 | Yes | ||
| 222 | SS18 | 18476 | -3.521 | -0.0291 | Yes | ||
| 223 | RIPK1 | 18527 | -3.741 | -0.0201 | Yes | ||
| 224 | CDYL | 18552 | -3.934 | -0.0092 | Yes | ||
| 225 | STAM | 18565 | -4.049 | 0.0028 | Yes |
