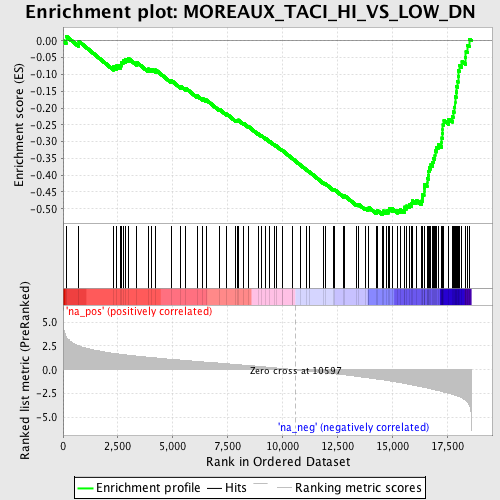
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MOREAUX_TACI_HI_VS_LOW_DN |
| Enrichment Score (ES) | -0.51712126 |
| Normalized Enrichment Score (NES) | -2.363092 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | GPR89A | 137 | 3.442 | 0.0141 | No | ||
| 2 | ERCC1 | 721 | 2.491 | -0.0018 | No | ||
| 3 | NNT | 2301 | 1.716 | -0.0764 | No | ||
| 4 | RUVBL2 | 2450 | 1.673 | -0.0739 | No | ||
| 5 | MRPL43 | 2610 | 1.633 | -0.0723 | No | ||
| 6 | MRPS12 | 2660 | 1.617 | -0.0648 | No | ||
| 7 | TRIB1 | 2735 | 1.594 | -0.0589 | No | ||
| 8 | MINPP1 | 2848 | 1.563 | -0.0551 | No | ||
| 9 | TTRAP | 2971 | 1.530 | -0.0522 | No | ||
| 10 | CYP2R1 | 3360 | 1.430 | -0.0642 | No | ||
| 11 | SLC33A1 | 3873 | 1.312 | -0.0836 | No | ||
| 12 | STAM2 | 4042 | 1.280 | -0.0847 | No | ||
| 13 | MYLIP | 4221 | 1.242 | -0.0866 | No | ||
| 14 | FIBP | 4921 | 1.093 | -0.1175 | No | ||
| 15 | PSMC4 | 5367 | 1.009 | -0.1352 | No | ||
| 16 | TIMM50 | 5588 | 0.966 | -0.1411 | No | ||
| 17 | TYMS | 6108 | 0.872 | -0.1637 | No | ||
| 18 | PPP2R5C | 6354 | 0.829 | -0.1717 | No | ||
| 19 | JTV1 | 6525 | 0.797 | -0.1759 | No | ||
| 20 | LAP3 | 7111 | 0.697 | -0.2032 | No | ||
| 21 | NDUFA13 | 7453 | 0.635 | -0.2176 | No | ||
| 22 | PAK1IP1 | 7870 | 0.554 | -0.2366 | No | ||
| 23 | IFT74 | 7964 | 0.537 | -0.2383 | No | ||
| 24 | BUB3 | 7978 | 0.535 | -0.2357 | No | ||
| 25 | SLC16A1 | 8235 | 0.486 | -0.2464 | No | ||
| 26 | MET | 8434 | 0.448 | -0.2543 | No | ||
| 27 | LSM4 | 8897 | 0.364 | -0.2770 | No | ||
| 28 | GEMIN5 | 9038 | 0.336 | -0.2825 | No | ||
| 29 | MCM2 | 9236 | 0.291 | -0.2913 | No | ||
| 30 | CCDC5 | 9424 | 0.249 | -0.2999 | No | ||
| 31 | LSM5 | 9635 | 0.202 | -0.3099 | No | ||
| 32 | TMEM165 | 9706 | 0.187 | -0.3125 | No | ||
| 33 | THYN1 | 9981 | 0.131 | -0.3265 | No | ||
| 34 | MCM3AP | 9986 | 0.131 | -0.3259 | No | ||
| 35 | TERF1 | 10434 | 0.034 | -0.3499 | No | ||
| 36 | PAPOLA | 10461 | 0.030 | -0.3511 | No | ||
| 37 | PSMD8 | 10838 | -0.059 | -0.3710 | No | ||
| 38 | ABHD3 | 11099 | -0.114 | -0.3844 | No | ||
| 39 | DHX29 | 11214 | -0.142 | -0.3896 | No | ||
| 40 | NUDT5 | 11869 | -0.291 | -0.4231 | No | ||
| 41 | CCT2 | 11951 | -0.312 | -0.4256 | No | ||
| 42 | COMMD4 | 12335 | -0.404 | -0.4437 | No | ||
| 43 | TLE4 | 12360 | -0.409 | -0.4425 | No | ||
| 44 | CHEK1 | 12792 | -0.517 | -0.4625 | No | ||
| 45 | RNF7 | 12834 | -0.528 | -0.4614 | No | ||
| 46 | PDCD6 | 13376 | -0.685 | -0.4864 | No | ||
| 47 | RAN | 13479 | -0.712 | -0.4874 | No | ||
| 48 | SNRPD1 | 13770 | -0.803 | -0.4981 | No | ||
| 49 | CDC23 | 13900 | -0.834 | -0.4998 | No | ||
| 50 | MRPL19 | 13940 | -0.847 | -0.4967 | No | ||
| 51 | TATDN1 | 14291 | -0.962 | -0.5096 | No | ||
| 52 | ANAPC10 | 14310 | -0.967 | -0.5045 | No | ||
| 53 | CIRH1A | 14545 | -1.038 | -0.5106 | Yes | ||
| 54 | SHFM1 | 14593 | -1.056 | -0.5066 | Yes | ||
| 55 | EBNA1BP2 | 14723 | -1.106 | -0.5066 | Yes | ||
| 56 | CCT5 | 14839 | -1.145 | -0.5057 | Yes | ||
| 57 | EFTUD2 | 14861 | -1.156 | -0.4996 | Yes | ||
| 58 | EIF3S4 | 15010 | -1.211 | -0.5000 | Yes | ||
| 59 | ACTR6 | 15251 | -1.309 | -0.5048 | Yes | ||
| 60 | RACGAP1 | 15365 | -1.348 | -0.5025 | Yes | ||
| 61 | PFDN4 | 15546 | -1.420 | -0.5033 | Yes | ||
| 62 | C1QBP | 15555 | -1.426 | -0.4948 | Yes | ||
| 63 | SF3B2 | 15670 | -1.478 | -0.4917 | Yes | ||
| 64 | UPF3A | 15780 | -1.524 | -0.4881 | Yes | ||
| 65 | HNRPAB | 15870 | -1.565 | -0.4831 | Yes | ||
| 66 | CHCHD1 | 15922 | -1.587 | -0.4760 | Yes | ||
| 67 | MINA | 16087 | -1.665 | -0.4744 | Yes | ||
| 68 | ORC5L | 16341 | -1.784 | -0.4769 | Yes | ||
| 69 | PAIP1 | 16376 | -1.799 | -0.4675 | Yes | ||
| 70 | ASF1A | 16393 | -1.808 | -0.4571 | Yes | ||
| 71 | EZH2 | 16450 | -1.831 | -0.4486 | Yes | ||
| 72 | ACTL6A | 16453 | -1.832 | -0.4373 | Yes | ||
| 73 | AASDHPPT | 16476 | -1.842 | -0.4270 | Yes | ||
| 74 | BARD1 | 16590 | -1.904 | -0.4212 | Yes | ||
| 75 | PCID1 | 16592 | -1.906 | -0.4093 | Yes | ||
| 76 | DARS | 16640 | -1.928 | -0.3998 | Yes | ||
| 77 | SMARCA4 | 16653 | -1.934 | -0.3883 | Yes | ||
| 78 | MATR3 | 16687 | -1.952 | -0.3779 | Yes | ||
| 79 | BTBD1 | 16763 | -1.998 | -0.3695 | Yes | ||
| 80 | PAPD4 | 16842 | -2.038 | -0.3609 | Yes | ||
| 81 | RNF111 | 16875 | -2.055 | -0.3498 | Yes | ||
| 82 | SGOL2 | 16947 | -2.091 | -0.3406 | Yes | ||
| 83 | TSHZ1 | 16951 | -2.092 | -0.3277 | Yes | ||
| 84 | PPP1R12A | 17014 | -2.128 | -0.3177 | Yes | ||
| 85 | KEAP1 | 17094 | -2.168 | -0.3084 | Yes | ||
| 86 | FEM1B | 17259 | -2.264 | -0.3031 | Yes | ||
| 87 | PRKDC | 17266 | -2.267 | -0.2893 | Yes | ||
| 88 | FANCL | 17299 | -2.290 | -0.2767 | Yes | ||
| 89 | IREB2 | 17308 | -2.294 | -0.2628 | Yes | ||
| 90 | SNX19 | 17313 | -2.297 | -0.2486 | Yes | ||
| 91 | ANKRD49 | 17358 | -2.325 | -0.2365 | Yes | ||
| 92 | KLF13 | 17576 | -2.454 | -0.2329 | Yes | ||
| 93 | NASP | 17744 | -2.569 | -0.2258 | Yes | ||
| 94 | SPATA5L1 | 17778 | -2.586 | -0.2114 | Yes | ||
| 95 | IARS | 17819 | -2.617 | -0.1972 | Yes | ||
| 96 | KRIT1 | 17861 | -2.657 | -0.1828 | Yes | ||
| 97 | PSMA3 | 17865 | -2.659 | -0.1664 | Yes | ||
| 98 | NMT1 | 17913 | -2.693 | -0.1521 | Yes | ||
| 99 | DDX31 | 17930 | -2.704 | -0.1360 | Yes | ||
| 100 | IFT57 | 17971 | -2.741 | -0.1210 | Yes | ||
| 101 | DDX1 | 18002 | -2.768 | -0.1054 | Yes | ||
| 102 | ILF3 | 18021 | -2.785 | -0.0889 | Yes | ||
| 103 | RSF1 | 18074 | -2.833 | -0.0740 | Yes | ||
| 104 | MRPL50 | 18179 | -2.945 | -0.0612 | Yes | ||
| 105 | DDX18 | 18344 | -3.197 | -0.0501 | Yes | ||
| 106 | IK | 18361 | -3.235 | -0.0307 | Yes | ||
| 107 | ANKRD17 | 18423 | -3.382 | -0.0129 | Yes | ||
| 108 | SPAST | 18522 | -3.717 | 0.0051 | Yes |
