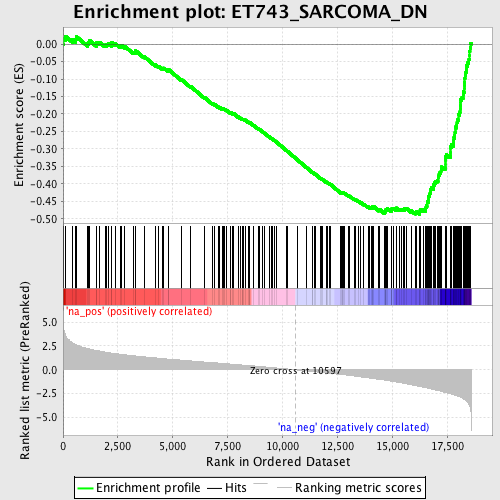
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ET743_SARCOMA_DN |
| Enrichment Score (ES) | -0.48785374 |
| Normalized Enrichment Score (NES) | -2.47576 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | WNT5B | 10 | 4.992 | 0.0157 | No | ||
| 2 | KIAA0859 | 96 | 3.643 | 0.0229 | No | ||
| 3 | HIST1H4C | 425 | 2.815 | 0.0142 | No | ||
| 4 | DLG5 | 586 | 2.628 | 0.0141 | No | ||
| 5 | LHFPL2 | 613 | 2.599 | 0.0211 | No | ||
| 6 | THRAP3 | 1116 | 2.213 | 0.0010 | No | ||
| 7 | EGFR | 1159 | 2.187 | 0.0059 | No | ||
| 8 | HIST1H4B | 1214 | 2.159 | 0.0099 | No | ||
| 9 | FYN | 1528 | 2.012 | -0.0005 | No | ||
| 10 | ZDHHC16 | 1536 | 2.010 | 0.0056 | No | ||
| 11 | FADD | 1663 | 1.958 | 0.0051 | No | ||
| 12 | SNIP1 | 1911 | 1.857 | -0.0022 | No | ||
| 13 | LUZP1 | 1991 | 1.827 | -0.0006 | No | ||
| 14 | SLC2A1 | 2059 | 1.804 | 0.0016 | No | ||
| 15 | HSPE1 | 2202 | 1.750 | -0.0004 | No | ||
| 16 | CRK | 2219 | 1.743 | 0.0044 | No | ||
| 17 | RBM14 | 2381 | 1.692 | 0.0012 | No | ||
| 18 | PAFAH2 | 2596 | 1.636 | -0.0051 | No | ||
| 19 | BAG2 | 2682 | 1.610 | -0.0045 | No | ||
| 20 | BLZF1 | 2812 | 1.574 | -0.0064 | No | ||
| 21 | NUFIP1 | 3206 | 1.467 | -0.0230 | No | ||
| 22 | SIRPA | 3291 | 1.445 | -0.0229 | No | ||
| 23 | PLK1 | 3309 | 1.442 | -0.0191 | No | ||
| 24 | CYHR1 | 3692 | 1.355 | -0.0354 | No | ||
| 25 | TIAM2 | 4204 | 1.246 | -0.0591 | No | ||
| 26 | ARFGAP1 | 4362 | 1.208 | -0.0637 | No | ||
| 27 | HS3ST3B1 | 4544 | 1.166 | -0.0698 | No | ||
| 28 | HSPA1L | 4589 | 1.158 | -0.0684 | No | ||
| 29 | PDGFB | 4781 | 1.119 | -0.0751 | No | ||
| 30 | B4GALT5 | 4805 | 1.115 | -0.0728 | No | ||
| 31 | PANX1 | 5402 | 1.002 | -0.1019 | No | ||
| 32 | MAP3K4 | 5813 | 0.926 | -0.1211 | No | ||
| 33 | CDK7 | 6460 | 0.809 | -0.1535 | No | ||
| 34 | RAB5A | 6825 | 0.738 | -0.1709 | No | ||
| 35 | JAG1 | 6897 | 0.728 | -0.1724 | No | ||
| 36 | IPO7 | 7076 | 0.702 | -0.1798 | No | ||
| 37 | KIF11 | 7145 | 0.690 | -0.1812 | No | ||
| 38 | USP3 | 7263 | 0.669 | -0.1854 | No | ||
| 39 | CXXC5 | 7290 | 0.666 | -0.1847 | No | ||
| 40 | BIRC6 | 7356 | 0.654 | -0.1861 | No | ||
| 41 | CDC42BPA | 7458 | 0.634 | -0.1895 | No | ||
| 42 | MID1IP1 | 7621 | 0.601 | -0.1963 | No | ||
| 43 | CTGF | 7634 | 0.599 | -0.1950 | No | ||
| 44 | CFLAR | 7738 | 0.579 | -0.1987 | No | ||
| 45 | TDG | 7781 | 0.570 | -0.1992 | No | ||
| 46 | BUB3 | 7978 | 0.535 | -0.2081 | No | ||
| 47 | EPS15 | 8074 | 0.516 | -0.2116 | No | ||
| 48 | PSMA5 | 8161 | 0.498 | -0.2146 | No | ||
| 49 | CLCN7 | 8194 | 0.494 | -0.2147 | No | ||
| 50 | BTG1 | 8242 | 0.484 | -0.2157 | No | ||
| 51 | XRCC5 | 8298 | 0.473 | -0.2172 | No | ||
| 52 | FOSL1 | 8432 | 0.449 | -0.2229 | No | ||
| 53 | EPS8 | 8493 | 0.436 | -0.2248 | No | ||
| 54 | ARPC5L | 8502 | 0.435 | -0.2238 | No | ||
| 55 | ADSL | 8696 | 0.399 | -0.2330 | No | ||
| 56 | ACSL3 | 8893 | 0.365 | -0.2424 | No | ||
| 57 | MFAP1 | 8942 | 0.354 | -0.2439 | No | ||
| 58 | OSBP | 9081 | 0.327 | -0.2503 | No | ||
| 59 | MTMR4 | 9191 | 0.300 | -0.2553 | No | ||
| 60 | FZD1 | 9395 | 0.257 | -0.2655 | No | ||
| 61 | UBN1 | 9494 | 0.234 | -0.2700 | No | ||
| 62 | NPAS2 | 9550 | 0.222 | -0.2723 | No | ||
| 63 | RCC1 | 9622 | 0.206 | -0.2755 | No | ||
| 64 | CRSP6 | 9743 | 0.180 | -0.2814 | No | ||
| 65 | BUB1 | 10189 | 0.087 | -0.3053 | No | ||
| 66 | MCM3 | 10237 | 0.078 | -0.3076 | No | ||
| 67 | UBE2C | 10662 | -0.017 | -0.3305 | No | ||
| 68 | ARHGEF2 | 11096 | -0.114 | -0.3537 | No | ||
| 69 | SFRS9 | 11352 | -0.172 | -0.3669 | No | ||
| 70 | NMI | 11384 | -0.180 | -0.3680 | No | ||
| 71 | FBXL7 | 11441 | -0.197 | -0.3704 | No | ||
| 72 | VEGFC | 11480 | -0.206 | -0.3718 | No | ||
| 73 | PSMC2 | 11717 | -0.255 | -0.3838 | No | ||
| 74 | CORO1C | 11788 | -0.273 | -0.3867 | No | ||
| 75 | BCL2L1 | 11807 | -0.276 | -0.3868 | No | ||
| 76 | TRIP4 | 11989 | -0.322 | -0.3956 | No | ||
| 77 | GDI2 | 11992 | -0.322 | -0.3946 | No | ||
| 78 | PSMB1 | 12039 | -0.334 | -0.3961 | No | ||
| 79 | MAPKAPK5 | 12126 | -0.351 | -0.3996 | No | ||
| 80 | TRIM24 | 12170 | -0.365 | -0.4007 | No | ||
| 81 | ARFGEF2 | 12660 | -0.487 | -0.4257 | No | ||
| 82 | MTIF2 | 12710 | -0.497 | -0.4267 | No | ||
| 83 | SPRY2 | 12721 | -0.500 | -0.4257 | No | ||
| 84 | TARDBP | 12728 | -0.503 | -0.4244 | No | ||
| 85 | YWHAE | 12773 | -0.513 | -0.4251 | No | ||
| 86 | PRIM2A | 12842 | -0.531 | -0.4270 | No | ||
| 87 | RCOR1 | 13002 | -0.575 | -0.4338 | No | ||
| 88 | MRPL15 | 13057 | -0.592 | -0.4348 | No | ||
| 89 | CSNK2A1 | 13267 | -0.649 | -0.4441 | No | ||
| 90 | PLAG1 | 13330 | -0.670 | -0.4452 | No | ||
| 91 | GGPS1 | 13447 | -0.704 | -0.4493 | No | ||
| 92 | PSME3 | 13560 | -0.736 | -0.4529 | No | ||
| 93 | CRLF3 | 13705 | -0.782 | -0.4582 | No | ||
| 94 | CDC23 | 13900 | -0.834 | -0.4660 | No | ||
| 95 | FBXL3 | 13986 | -0.860 | -0.4679 | No | ||
| 96 | MRPS35 | 14036 | -0.878 | -0.4677 | No | ||
| 97 | GABPB2 | 14094 | -0.901 | -0.4678 | No | ||
| 98 | MYC | 14099 | -0.902 | -0.4651 | No | ||
| 99 | RAD23B | 14126 | -0.911 | -0.4636 | No | ||
| 100 | CDK5R1 | 14394 | -0.995 | -0.4748 | No | ||
| 101 | SSFA2 | 14406 | -0.999 | -0.4722 | No | ||
| 102 | PPP1CC | 14643 | -1.074 | -0.4815 | No | ||
| 103 | ERCC3 | 14676 | -1.088 | -0.4797 | No | ||
| 104 | STK17B | 14686 | -1.092 | -0.4767 | No | ||
| 105 | ATF1 | 14712 | -1.102 | -0.4744 | No | ||
| 106 | ZDHHC5 | 14758 | -1.119 | -0.4733 | No | ||
| 107 | WEE1 | 14784 | -1.131 | -0.4709 | No | ||
| 108 | RBM27 | 14958 | -1.190 | -0.4765 | No | ||
| 109 | PTPRK | 14983 | -1.203 | -0.4739 | No | ||
| 110 | EIF4B | 14986 | -1.203 | -0.4701 | No | ||
| 111 | AURKA | 15058 | -1.229 | -0.4699 | No | ||
| 112 | PSMC6 | 15180 | -1.279 | -0.4723 | No | ||
| 113 | FAF1 | 15198 | -1.285 | -0.4691 | No | ||
| 114 | EML4 | 15341 | -1.339 | -0.4725 | No | ||
| 115 | CDH11 | 15436 | -1.378 | -0.4731 | No | ||
| 116 | MAPK14 | 15512 | -1.409 | -0.4726 | No | ||
| 117 | TCERG1 | 15569 | -1.433 | -0.4710 | No | ||
| 118 | TMPO | 15636 | -1.460 | -0.4698 | No | ||
| 119 | USP12 | 15860 | -1.564 | -0.4768 | No | ||
| 120 | SAMD4A | 16042 | -1.646 | -0.4813 | No | ||
| 121 | PRKD3 | 16089 | -1.667 | -0.4784 | No | ||
| 122 | FNDC3A | 16264 | -1.741 | -0.4822 | Yes | ||
| 123 | ATG5 | 16265 | -1.742 | -0.4765 | Yes | ||
| 124 | CDC25C | 16298 | -1.762 | -0.4726 | Yes | ||
| 125 | SUPV3L1 | 16436 | -1.826 | -0.4741 | Yes | ||
| 126 | HSPH1 | 16524 | -1.868 | -0.4727 | Yes | ||
| 127 | NCOA6 | 16535 | -1.874 | -0.4672 | Yes | ||
| 128 | JMJD2B | 16559 | -1.884 | -0.4623 | Yes | ||
| 129 | TNFSF5IP1 | 16588 | -1.902 | -0.4577 | Yes | ||
| 130 | OSBPL9 | 16589 | -1.904 | -0.4515 | Yes | ||
| 131 | TOP2A | 16650 | -1.933 | -0.4485 | Yes | ||
| 132 | UBE2D2 | 16660 | -1.936 | -0.4427 | Yes | ||
| 133 | POLD3 | 16664 | -1.938 | -0.4365 | Yes | ||
| 134 | PPP2R5E | 16688 | -1.953 | -0.4314 | Yes | ||
| 135 | XBP1 | 16718 | -1.969 | -0.4266 | Yes | ||
| 136 | RAP1B | 16745 | -1.987 | -0.4216 | Yes | ||
| 137 | MAT2A | 16755 | -1.992 | -0.4156 | Yes | ||
| 138 | DCK | 16792 | -2.015 | -0.4110 | Yes | ||
| 139 | SIRT1 | 16877 | -2.056 | -0.4089 | Yes | ||
| 140 | RAB6A | 16886 | -2.059 | -0.4026 | Yes | ||
| 141 | USP8 | 16914 | -2.072 | -0.3974 | Yes | ||
| 142 | PHF6 | 16970 | -2.099 | -0.3935 | Yes | ||
| 143 | JMJD1C | 17044 | -2.143 | -0.3905 | Yes | ||
| 144 | UBE2D3 | 17090 | -2.167 | -0.3859 | Yes | ||
| 145 | PCNA | 17091 | -2.167 | -0.3789 | Yes | ||
| 146 | SON | 17130 | -2.195 | -0.3738 | Yes | ||
| 147 | MAP3K7 | 17171 | -2.215 | -0.3688 | Yes | ||
| 148 | PJA2 | 17221 | -2.243 | -0.3642 | Yes | ||
| 149 | NBN | 17226 | -2.246 | -0.3571 | Yes | ||
| 150 | GOPC | 17260 | -2.265 | -0.3516 | Yes | ||
| 151 | RABGGTB | 17411 | -2.356 | -0.3520 | Yes | ||
| 152 | MCTS1 | 17417 | -2.359 | -0.3447 | Yes | ||
| 153 | DDX21 | 17421 | -2.360 | -0.3372 | Yes | ||
| 154 | RNF14 | 17424 | -2.362 | -0.3296 | Yes | ||
| 155 | CDC5L | 17441 | -2.373 | -0.3228 | Yes | ||
| 156 | RBM17 | 17470 | -2.389 | -0.3165 | Yes | ||
| 157 | GTF2H1 | 17644 | -2.502 | -0.3178 | Yes | ||
| 158 | EIF3S10 | 17651 | -2.505 | -0.3100 | Yes | ||
| 159 | PSMA4 | 17660 | -2.511 | -0.3023 | Yes | ||
| 160 | SRRM2 | 17662 | -2.511 | -0.2942 | Yes | ||
| 161 | FNTA | 17695 | -2.534 | -0.2877 | Yes | ||
| 162 | HSPA8 | 17780 | -2.588 | -0.2838 | Yes | ||
| 163 | GTF2H2 | 17799 | -2.602 | -0.2763 | Yes | ||
| 164 | AOF2 | 17811 | -2.611 | -0.2685 | Yes | ||
| 165 | PKN2 | 17845 | -2.641 | -0.2617 | Yes | ||
| 166 | TBK1 | 17859 | -2.654 | -0.2538 | Yes | ||
| 167 | SFRS3 | 17871 | -2.665 | -0.2457 | Yes | ||
| 168 | CCT4 | 17874 | -2.665 | -0.2372 | Yes | ||
| 169 | HDAC2 | 17934 | -2.708 | -0.2316 | Yes | ||
| 170 | OGT | 17951 | -2.718 | -0.2236 | Yes | ||
| 171 | RPP14 | 17991 | -2.757 | -0.2168 | Yes | ||
| 172 | PRKACB | 18025 | -2.788 | -0.2095 | Yes | ||
| 173 | SLC25A17 | 18026 | -2.789 | -0.2005 | Yes | ||
| 174 | RABGAP1 | 18084 | -2.842 | -0.1943 | Yes | ||
| 175 | STAU1 | 18093 | -2.851 | -0.1855 | Yes | ||
| 176 | MRPL1 | 18103 | -2.863 | -0.1767 | Yes | ||
| 177 | TOP1 | 18114 | -2.872 | -0.1679 | Yes | ||
| 178 | SMARCA5 | 18120 | -2.882 | -0.1588 | Yes | ||
| 179 | RAB1A | 18177 | -2.944 | -0.1523 | Yes | ||
| 180 | CCNA2 | 18230 | -3.023 | -0.1453 | Yes | ||
| 181 | LBR | 18241 | -3.037 | -0.1360 | Yes | ||
| 182 | TXNRD1 | 18280 | -3.104 | -0.1280 | Yes | ||
| 183 | SRPK2 | 18286 | -3.115 | -0.1181 | Yes | ||
| 184 | PSMD14 | 18296 | -3.126 | -0.1085 | Yes | ||
| 185 | EP400 | 18309 | -3.142 | -0.0989 | Yes | ||
| 186 | CCNH | 18320 | -3.158 | -0.0892 | Yes | ||
| 187 | DNAJA2 | 18353 | -3.221 | -0.0805 | Yes | ||
| 188 | BIRC2 | 18371 | -3.248 | -0.0709 | Yes | ||
| 189 | NET1 | 18397 | -3.291 | -0.0615 | Yes | ||
| 190 | ELOVL6 | 18445 | -3.425 | -0.0530 | Yes | ||
| 191 | GTF2B | 18495 | -3.569 | -0.0440 | Yes | ||
| 192 | DDX20 | 18519 | -3.704 | -0.0332 | Yes | ||
| 193 | RIPK1 | 18527 | -3.741 | -0.0215 | Yes | ||
| 194 | ANLN | 18546 | -3.873 | -0.0099 | Yes | ||
| 195 | SLC4A1AP | 18579 | -4.194 | 0.0020 | Yes |
