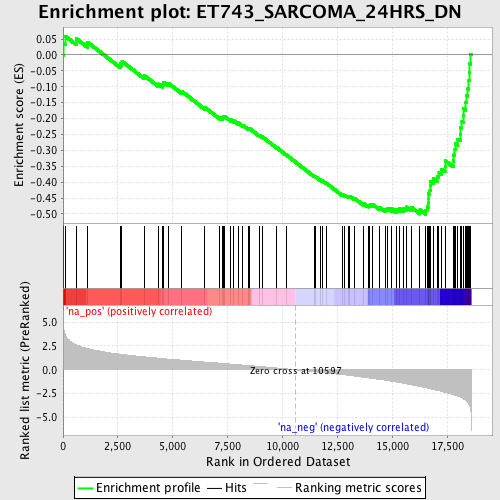
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ET743_SARCOMA_24HRS_DN |
| Enrichment Score (ES) | -0.50177705 |
| Normalized Enrichment Score (NES) | -2.2395988 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.2163287E-4 |
| FWER p-Value | 0.0070 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | WNT5B | 10 | 4.992 | 0.0367 | No | ||
| 2 | KIAA0859 | 96 | 3.643 | 0.0594 | No | ||
| 3 | LHFPL2 | 613 | 2.599 | 0.0509 | No | ||
| 4 | THRAP3 | 1116 | 2.213 | 0.0404 | No | ||
| 5 | PAFAH2 | 2596 | 1.636 | -0.0272 | No | ||
| 6 | BAG2 | 2682 | 1.610 | -0.0198 | No | ||
| 7 | CYHR1 | 3692 | 1.355 | -0.0641 | No | ||
| 8 | ARFGAP1 | 4362 | 1.208 | -0.0912 | No | ||
| 9 | HS3ST3B1 | 4544 | 1.166 | -0.0922 | No | ||
| 10 | HSPA1L | 4589 | 1.158 | -0.0860 | No | ||
| 11 | B4GALT5 | 4805 | 1.115 | -0.0892 | No | ||
| 12 | PANX1 | 5402 | 1.002 | -0.1139 | No | ||
| 13 | CDK7 | 6460 | 0.809 | -0.1649 | No | ||
| 14 | KIF11 | 7145 | 0.690 | -0.1966 | No | ||
| 15 | USP3 | 7263 | 0.669 | -0.1980 | No | ||
| 16 | CXXC5 | 7290 | 0.666 | -0.1944 | No | ||
| 17 | BIRC6 | 7356 | 0.654 | -0.1930 | No | ||
| 18 | CTGF | 7634 | 0.599 | -0.2035 | No | ||
| 19 | TDG | 7781 | 0.570 | -0.2071 | No | ||
| 20 | BUB3 | 7978 | 0.535 | -0.2137 | No | ||
| 21 | CLCN7 | 8194 | 0.494 | -0.2216 | No | ||
| 22 | FOSL1 | 8432 | 0.449 | -0.2310 | No | ||
| 23 | EPS8 | 8493 | 0.436 | -0.2310 | No | ||
| 24 | MFAP1 | 8942 | 0.354 | -0.2525 | No | ||
| 25 | OSBP | 9081 | 0.327 | -0.2576 | No | ||
| 26 | CRSP6 | 9743 | 0.180 | -0.2919 | No | ||
| 27 | BUB1 | 10189 | 0.087 | -0.3152 | No | ||
| 28 | FBXL7 | 11441 | -0.197 | -0.3813 | No | ||
| 29 | VEGFC | 11480 | -0.206 | -0.3818 | No | ||
| 30 | PSMC2 | 11717 | -0.255 | -0.3926 | No | ||
| 31 | BCL2L1 | 11807 | -0.276 | -0.3953 | No | ||
| 32 | TRIP4 | 11989 | -0.322 | -0.4027 | No | ||
| 33 | TARDBP | 12728 | -0.503 | -0.4388 | No | ||
| 34 | PRIM2A | 12842 | -0.531 | -0.4409 | No | ||
| 35 | RCOR1 | 13002 | -0.575 | -0.4452 | No | ||
| 36 | MRPL15 | 13057 | -0.592 | -0.4437 | No | ||
| 37 | CSNK2A1 | 13267 | -0.649 | -0.4501 | No | ||
| 38 | CRLF3 | 13705 | -0.782 | -0.4678 | No | ||
| 39 | CDC23 | 13900 | -0.834 | -0.4721 | No | ||
| 40 | FBXL3 | 13986 | -0.860 | -0.4702 | No | ||
| 41 | GABPB2 | 14094 | -0.901 | -0.4693 | No | ||
| 42 | SSFA2 | 14406 | -0.999 | -0.4786 | No | ||
| 43 | STK17B | 14686 | -1.092 | -0.4855 | No | ||
| 44 | WEE1 | 14784 | -1.131 | -0.4823 | No | ||
| 45 | RBM27 | 14958 | -1.190 | -0.4827 | No | ||
| 46 | PSMC6 | 15180 | -1.279 | -0.4851 | No | ||
| 47 | EML4 | 15341 | -1.339 | -0.4837 | No | ||
| 48 | MAPK14 | 15512 | -1.409 | -0.4824 | No | ||
| 49 | TMPO | 15636 | -1.460 | -0.4781 | No | ||
| 50 | USP12 | 15860 | -1.564 | -0.4785 | No | ||
| 51 | ATG5 | 16265 | -1.742 | -0.4873 | No | ||
| 52 | NCOA6 | 16535 | -1.874 | -0.4878 | Yes | ||
| 53 | TNFSF5IP1 | 16588 | -1.902 | -0.4764 | Yes | ||
| 54 | TOP2A | 16650 | -1.933 | -0.4652 | Yes | ||
| 55 | UBE2D2 | 16660 | -1.936 | -0.4513 | Yes | ||
| 56 | POLD3 | 16664 | -1.938 | -0.4370 | Yes | ||
| 57 | XBP1 | 16718 | -1.969 | -0.4251 | Yes | ||
| 58 | RAP1B | 16745 | -1.987 | -0.4117 | Yes | ||
| 59 | MAT2A | 16755 | -1.992 | -0.3973 | Yes | ||
| 60 | SIRT1 | 16877 | -2.056 | -0.3885 | Yes | ||
| 61 | JMJD1C | 17044 | -2.143 | -0.3814 | Yes | ||
| 62 | SON | 17130 | -2.195 | -0.3696 | Yes | ||
| 63 | GOPC | 17260 | -2.265 | -0.3597 | Yes | ||
| 64 | RABGGTB | 17411 | -2.356 | -0.3502 | Yes | ||
| 65 | RNF14 | 17424 | -2.362 | -0.3332 | Yes | ||
| 66 | HSPA8 | 17780 | -2.588 | -0.3330 | Yes | ||
| 67 | GTF2H2 | 17799 | -2.602 | -0.3146 | Yes | ||
| 68 | TBK1 | 17859 | -2.654 | -0.2979 | Yes | ||
| 69 | SFRS3 | 17871 | -2.665 | -0.2786 | Yes | ||
| 70 | RPP14 | 17991 | -2.757 | -0.2644 | Yes | ||
| 71 | MRPL1 | 18103 | -2.863 | -0.2491 | Yes | ||
| 72 | TOP1 | 18114 | -2.872 | -0.2281 | Yes | ||
| 73 | RAB1A | 18177 | -2.944 | -0.2095 | Yes | ||
| 74 | CCNA2 | 18230 | -3.023 | -0.1897 | Yes | ||
| 75 | LBR | 18241 | -3.037 | -0.1676 | Yes | ||
| 76 | DNAJA2 | 18353 | -3.221 | -0.1495 | Yes | ||
| 77 | BIRC2 | 18371 | -3.248 | -0.1262 | Yes | ||
| 78 | ELOVL6 | 18445 | -3.425 | -0.1046 | Yes | ||
| 79 | GTF2B | 18495 | -3.569 | -0.0805 | Yes | ||
| 80 | DDX20 | 18519 | -3.704 | -0.0541 | Yes | ||
| 81 | RIPK1 | 18527 | -3.741 | -0.0266 | Yes | ||
| 82 | SLC4A1AP | 18579 | -4.194 | 0.0020 | Yes |
