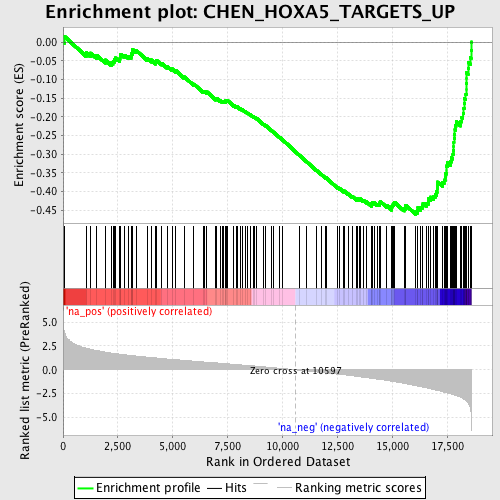
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_wtNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CHEN_HOXA5_TARGETS_UP |
| Enrichment Score (ES) | -0.46103323 |
| Normalized Enrichment Score (NES) | -2.26549 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.3126152E-4 |
| FWER p-Value | 0.0060 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MAFF | 47 | 4.115 | 0.0161 | No | ||
| 2 | TBX3 | 1050 | 2.255 | -0.0279 | No | ||
| 3 | CEBPB | 1251 | 2.138 | -0.0290 | No | ||
| 4 | TUFT1 | 1531 | 2.012 | -0.0349 | No | ||
| 5 | SCG5 | 1922 | 1.853 | -0.0476 | No | ||
| 6 | EGR1 | 2211 | 1.746 | -0.0553 | No | ||
| 7 | CREB5 | 2274 | 1.724 | -0.0508 | No | ||
| 8 | DUSP6 | 2326 | 1.706 | -0.0458 | No | ||
| 9 | RELB | 2399 | 1.686 | -0.0421 | No | ||
| 10 | GAD1 | 2588 | 1.639 | -0.0448 | No | ||
| 11 | IRF7 | 2594 | 1.637 | -0.0376 | No | ||
| 12 | POLR3F | 2624 | 1.629 | -0.0318 | No | ||
| 13 | TSC22D1 | 2814 | 1.574 | -0.0349 | No | ||
| 14 | LIF | 2994 | 1.523 | -0.0377 | No | ||
| 15 | WAC | 3097 | 1.498 | -0.0364 | No | ||
| 16 | IFI16 | 3128 | 1.490 | -0.0312 | No | ||
| 17 | CARS | 3156 | 1.480 | -0.0260 | No | ||
| 18 | TNFRSF9 | 3165 | 1.477 | -0.0197 | No | ||
| 19 | SEC31A | 3327 | 1.438 | -0.0219 | No | ||
| 20 | PSPC1 | 3832 | 1.322 | -0.0432 | No | ||
| 21 | HLX1 | 4021 | 1.284 | -0.0475 | No | ||
| 22 | SOX30 | 4229 | 1.239 | -0.0531 | No | ||
| 23 | DAAM1 | 4252 | 1.235 | -0.0487 | No | ||
| 24 | TPTE | 4484 | 1.181 | -0.0558 | No | ||
| 25 | DSCR1 | 4742 | 1.127 | -0.0646 | No | ||
| 26 | GADD45B | 4962 | 1.087 | -0.0715 | No | ||
| 27 | FOSL2 | 5125 | 1.056 | -0.0755 | No | ||
| 28 | TRPC1 | 5530 | 0.977 | -0.0929 | No | ||
| 29 | NFKBIA | 5957 | 0.899 | -0.1119 | No | ||
| 30 | SUHW4 | 6382 | 0.823 | -0.1311 | No | ||
| 31 | CDK7 | 6460 | 0.809 | -0.1316 | No | ||
| 32 | BDKRB1 | 6556 | 0.792 | -0.1332 | No | ||
| 33 | PTBP2 | 6960 | 0.720 | -0.1517 | No | ||
| 34 | ATF3 | 7003 | 0.714 | -0.1507 | No | ||
| 35 | CREM | 7150 | 0.689 | -0.1555 | No | ||
| 36 | BACH1 | 7275 | 0.667 | -0.1592 | No | ||
| 37 | JUN | 7319 | 0.660 | -0.1585 | No | ||
| 38 | NSUN6 | 7396 | 0.646 | -0.1597 | No | ||
| 39 | HRH1 | 7400 | 0.645 | -0.1569 | No | ||
| 40 | DLX2 | 7443 | 0.638 | -0.1563 | No | ||
| 41 | TMEFF1 | 7508 | 0.624 | -0.1569 | No | ||
| 42 | TDG | 7781 | 0.570 | -0.1691 | No | ||
| 43 | STC2 | 7892 | 0.549 | -0.1725 | No | ||
| 44 | PTN | 7932 | 0.541 | -0.1722 | No | ||
| 45 | FOS | 8087 | 0.514 | -0.1782 | No | ||
| 46 | AGTPBP1 | 8157 | 0.499 | -0.1797 | No | ||
| 47 | JUNB | 8312 | 0.471 | -0.1859 | No | ||
| 48 | PRKRIP1 | 8418 | 0.452 | -0.1895 | No | ||
| 49 | IER2 | 8537 | 0.429 | -0.1939 | No | ||
| 50 | KIF18A | 8662 | 0.405 | -0.1988 | No | ||
| 51 | PTPN2 | 8723 | 0.395 | -0.2003 | No | ||
| 52 | CLEC4A | 8825 | 0.377 | -0.2040 | No | ||
| 53 | NFIL3 | 9133 | 0.315 | -0.2192 | No | ||
| 54 | TIPARP | 9234 | 0.291 | -0.2233 | No | ||
| 55 | TNFAIP3 | 9502 | 0.232 | -0.2367 | No | ||
| 56 | FBXO38 | 9606 | 0.211 | -0.2413 | No | ||
| 57 | CTH | 9873 | 0.152 | -0.2550 | No | ||
| 58 | KLF6 | 9982 | 0.131 | -0.2603 | No | ||
| 59 | KLF5 | 10788 | -0.049 | -0.3036 | No | ||
| 60 | ZNF228 | 11092 | -0.113 | -0.3195 | No | ||
| 61 | ARID4B | 11551 | -0.218 | -0.3433 | No | ||
| 62 | DUSP8 | 11775 | -0.269 | -0.3542 | No | ||
| 63 | CRY1 | 11974 | -0.318 | -0.3634 | No | ||
| 64 | CASP3 | 11982 | -0.320 | -0.3624 | No | ||
| 65 | NCOA3 | 12515 | -0.452 | -0.3891 | No | ||
| 66 | CENPC1 | 12604 | -0.474 | -0.3917 | No | ||
| 67 | IFRD1 | 12779 | -0.515 | -0.3988 | No | ||
| 68 | JOSD3 | 12811 | -0.523 | -0.3981 | No | ||
| 69 | F3 | 13007 | -0.576 | -0.4060 | No | ||
| 70 | ZNF35 | 13180 | -0.623 | -0.4125 | No | ||
| 71 | FOSB | 13389 | -0.687 | -0.4207 | No | ||
| 72 | SAT1 | 13411 | -0.694 | -0.4186 | No | ||
| 73 | KLF10 | 13524 | -0.725 | -0.4214 | No | ||
| 74 | BCL10 | 13545 | -0.732 | -0.4192 | No | ||
| 75 | RCHY1 | 13687 | -0.776 | -0.4233 | No | ||
| 76 | PCTK2 | 13824 | -0.817 | -0.4269 | No | ||
| 77 | NFKBIE | 14047 | -0.881 | -0.4349 | No | ||
| 78 | MARK3 | 14066 | -0.889 | -0.4319 | No | ||
| 79 | GEM | 14083 | -0.895 | -0.4287 | No | ||
| 80 | NRBF2 | 14181 | -0.927 | -0.4297 | No | ||
| 81 | EIF5 | 14339 | -0.978 | -0.4338 | No | ||
| 82 | RYBP | 14422 | -1.003 | -0.4337 | No | ||
| 83 | ASNS | 14438 | -1.008 | -0.4299 | No | ||
| 84 | PTGS2 | 14448 | -1.011 | -0.4258 | No | ||
| 85 | RBM15 | 14754 | -1.117 | -0.4372 | No | ||
| 86 | CAB39 | 14980 | -1.201 | -0.4440 | No | ||
| 87 | SNAI2 | 14981 | -1.201 | -0.4385 | No | ||
| 88 | PMAIP1 | 15030 | -1.217 | -0.4356 | No | ||
| 89 | CHD9 | 15073 | -1.235 | -0.4322 | No | ||
| 90 | BIRC3 | 15101 | -1.247 | -0.4280 | No | ||
| 91 | ARNTL | 15540 | -1.419 | -0.4453 | No | ||
| 92 | SNX5 | 15609 | -1.449 | -0.4424 | No | ||
| 93 | ETS2 | 15613 | -1.451 | -0.4360 | No | ||
| 94 | ZNF3 | 16077 | -1.660 | -0.4535 | Yes | ||
| 95 | CCNT2 | 16150 | -1.693 | -0.4497 | Yes | ||
| 96 | DPP8 | 16171 | -1.700 | -0.4431 | Yes | ||
| 97 | NEDD9 | 16293 | -1.757 | -0.4416 | Yes | ||
| 98 | RBM5 | 16365 | -1.792 | -0.4373 | Yes | ||
| 99 | AKAP10 | 16400 | -1.812 | -0.4310 | Yes | ||
| 100 | ING3 | 16548 | -1.878 | -0.4304 | Yes | ||
| 101 | FNBP4 | 16638 | -1.928 | -0.4264 | Yes | ||
| 102 | LIG4 | 16666 | -1.938 | -0.4191 | Yes | ||
| 103 | ARIH1 | 16744 | -1.986 | -0.4142 | Yes | ||
| 104 | RNF111 | 16875 | -2.055 | -0.4120 | Yes | ||
| 105 | FEM1C | 16955 | -2.094 | -0.4067 | Yes | ||
| 106 | PPP1R12A | 17014 | -2.128 | -0.4002 | Yes | ||
| 107 | JMJD1C | 17044 | -2.143 | -0.3920 | Yes | ||
| 108 | RBBP6 | 17062 | -2.151 | -0.3832 | Yes | ||
| 109 | TNFRSF10B | 17075 | -2.158 | -0.3740 | Yes | ||
| 110 | NUPL1 | 17287 | -2.281 | -0.3751 | Yes | ||
| 111 | INTS6 | 17365 | -2.327 | -0.3687 | Yes | ||
| 112 | RABGGTB | 17411 | -2.356 | -0.3604 | Yes | ||
| 113 | CCNL1 | 17428 | -2.365 | -0.3506 | Yes | ||
| 114 | RBM39 | 17472 | -2.390 | -0.3420 | Yes | ||
| 115 | CNOT4 | 17473 | -2.390 | -0.3312 | Yes | ||
| 116 | ZZZ3 | 17512 | -2.418 | -0.3223 | Yes | ||
| 117 | TAF1A | 17668 | -2.517 | -0.3192 | Yes | ||
| 118 | SEC24A | 17697 | -2.534 | -0.3092 | Yes | ||
| 119 | STRN3 | 17765 | -2.578 | -0.3011 | Yes | ||
| 120 | SPATA5L1 | 17778 | -2.586 | -0.2900 | Yes | ||
| 121 | ZCCHC6 | 17804 | -2.604 | -0.2796 | Yes | ||
| 122 | NEK1 | 17809 | -2.610 | -0.2679 | Yes | ||
| 123 | TLK2 | 17816 | -2.616 | -0.2564 | Yes | ||
| 124 | PELI1 | 17846 | -2.641 | -0.2460 | Yes | ||
| 125 | TBK1 | 17859 | -2.654 | -0.2345 | Yes | ||
| 126 | MARCH7 | 17877 | -2.667 | -0.2234 | Yes | ||
| 127 | CNOT2 | 17907 | -2.690 | -0.2127 | Yes | ||
| 128 | ZRF1 | 18124 | -2.885 | -0.2113 | Yes | ||
| 129 | DMTF1 | 18169 | -2.940 | -0.2003 | Yes | ||
| 130 | CLK4 | 18225 | -3.018 | -0.1896 | Yes | ||
| 131 | SFPQ | 18263 | -3.078 | -0.1776 | Yes | ||
| 132 | SDCCAG1 | 18279 | -3.102 | -0.1643 | Yes | ||
| 133 | CLK1 | 18297 | -3.127 | -0.1511 | Yes | ||
| 134 | SNX16 | 18355 | -3.227 | -0.1395 | Yes | ||
| 135 | BIRC2 | 18371 | -3.248 | -0.1256 | Yes | ||
| 136 | RIPK2 | 18373 | -3.252 | -0.1108 | Yes | ||
| 137 | ZCCHC8 | 18390 | -3.281 | -0.0968 | Yes | ||
| 138 | TANK | 18393 | -3.284 | -0.0820 | Yes | ||
| 139 | SETDB1 | 18459 | -3.457 | -0.0698 | Yes | ||
| 140 | KIN | 18482 | -3.536 | -0.0549 | Yes | ||
| 141 | DCLRE1C | 18558 | -3.977 | -0.0409 | Yes | ||
| 142 | MCL1 | 18596 | -4.386 | -0.0230 | Yes | ||
| 143 | PRPF3 | 18608 | -5.298 | 0.0004 | Yes |
