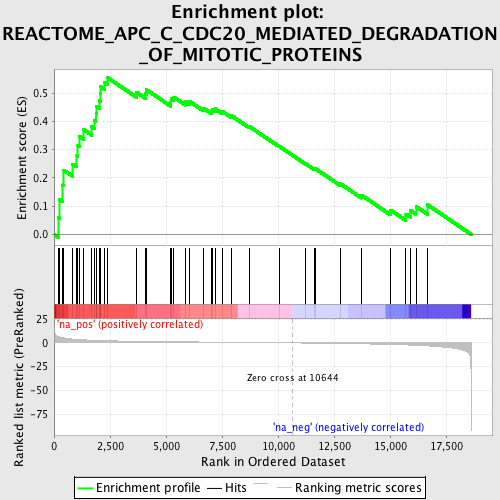
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_APC_C_CDC20_MEDIATED_DEGRADATION_OF_MITOTIC_PROTEINS |
| Enrichment Score (ES) | 0.5551074 |
| Normalized Enrichment Score (NES) | 1.9816606 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0037342815 |
| FWER p-Value | 0.075 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMC5 | 201 | 6.583 | 0.0592 | Yes | ||
| 2 | PSMB1 | 241 | 6.222 | 0.1233 | Yes | ||
| 3 | PSMB10 | 376 | 5.524 | 0.1749 | Yes | ||
| 4 | PSMB2 | 437 | 5.206 | 0.2271 | Yes | ||
| 5 | PSMD8 | 837 | 4.010 | 0.2482 | Yes | ||
| 6 | PSMC3 | 1017 | 3.685 | 0.2778 | Yes | ||
| 7 | PSMA7 | 1062 | 3.580 | 0.3135 | Yes | ||
| 8 | PSMB6 | 1130 | 3.476 | 0.3469 | Yes | ||
| 9 | PSMD9 | 1326 | 3.184 | 0.3703 | Yes | ||
| 10 | PSMA5 | 1679 | 2.796 | 0.3811 | Yes | ||
| 11 | PSME3 | 1801 | 2.682 | 0.4031 | Yes | ||
| 12 | PSMB3 | 1869 | 2.617 | 0.4274 | Yes | ||
| 13 | PSMB9 | 1884 | 2.601 | 0.4543 | Yes | ||
| 14 | CDC20 | 2036 | 2.471 | 0.4725 | Yes | ||
| 15 | PSMB5 | 2056 | 2.463 | 0.4977 | Yes | ||
| 16 | BUB1B | 2062 | 2.458 | 0.5235 | Yes | ||
| 17 | PSMC2 | 2265 | 2.319 | 0.5373 | Yes | ||
| 18 | PSMD3 | 2381 | 2.252 | 0.5551 | Yes | ||
| 19 | PSMD14 | 3686 | 1.665 | 0.5026 | No | ||
| 20 | PSMC6 | 4075 | 1.547 | 0.4982 | No | ||
| 21 | BUB3 | 4120 | 1.531 | 0.5121 | No | ||
| 22 | PSMC4 | 5210 | 1.245 | 0.4667 | No | ||
| 23 | PSMD11 | 5218 | 1.244 | 0.4796 | No | ||
| 24 | PSMD12 | 5339 | 1.212 | 0.4860 | No | ||
| 25 | PSMD5 | 5861 | 1.082 | 0.4695 | No | ||
| 26 | PSMC1 | 6024 | 1.047 | 0.4719 | No | ||
| 27 | PSMD2 | 6667 | 0.896 | 0.4469 | No | ||
| 28 | UBE2E1 | 7015 | 0.823 | 0.4369 | No | ||
| 29 | PSME1 | 7056 | 0.814 | 0.4435 | No | ||
| 30 | PTTG1 | 7198 | 0.783 | 0.4442 | No | ||
| 31 | PSMA1 | 7531 | 0.715 | 0.4339 | No | ||
| 32 | PSMA6 | 7916 | 0.633 | 0.4200 | No | ||
| 33 | PSMA4 | 8701 | 0.467 | 0.3827 | No | ||
| 34 | UBE2C | 10059 | 0.161 | 0.3114 | No | ||
| 35 | PSMA2 | 11218 | -0.151 | 0.2506 | No | ||
| 36 | MAD2L1 | 11628 | -0.269 | 0.2315 | No | ||
| 37 | PSMB4 | 11668 | -0.281 | 0.2324 | No | ||
| 38 | UBE2D1 | 12788 | -0.624 | 0.1788 | No | ||
| 39 | CDC16 | 13717 | -0.975 | 0.1392 | No | ||
| 40 | PSMD7 | 15027 | -1.615 | 0.0859 | No | ||
| 41 | PSMA3 | 15706 | -2.110 | 0.0718 | No | ||
| 42 | PSMD10 | 15899 | -2.279 | 0.0857 | No | ||
| 43 | CCNB1 | 16156 | -2.542 | 0.0990 | No | ||
| 44 | PSMD4 | 16665 | -3.138 | 0.1050 | No |