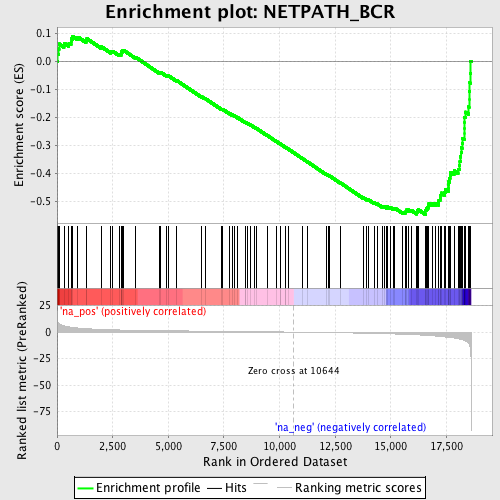
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | -0.54707766 |
| Normalized Enrichment Score (NES) | -1.9255245 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05241455 |
| FWER p-Value | 0.482 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BAX | 24 | 10.554 | 0.0252 | No | ||
| 2 | MAPKAPK2 | 58 | 8.591 | 0.0449 | No | ||
| 3 | CDK4 | 86 | 7.778 | 0.0630 | No | ||
| 4 | RPS6KA1 | 310 | 5.834 | 0.0655 | No | ||
| 5 | PIK3R2 | 532 | 4.858 | 0.0658 | No | ||
| 6 | GSK3A | 627 | 4.530 | 0.0721 | No | ||
| 7 | RPS6 | 634 | 4.515 | 0.0831 | No | ||
| 8 | CRK | 693 | 4.360 | 0.0909 | No | ||
| 9 | DOK1 | 933 | 3.839 | 0.0876 | No | ||
| 10 | BLK | 1301 | 3.219 | 0.0758 | No | ||
| 11 | ZAP70 | 1339 | 3.169 | 0.0817 | No | ||
| 12 | DOK3 | 1976 | 2.511 | 0.0537 | No | ||
| 13 | SHC1 | 2417 | 2.229 | 0.0355 | No | ||
| 14 | LAT2 | 2496 | 2.181 | 0.0367 | No | ||
| 15 | CD22 | 2798 | 2.018 | 0.0255 | No | ||
| 16 | FCGR2B | 2874 | 1.982 | 0.0264 | No | ||
| 17 | CD72 | 2875 | 1.982 | 0.0314 | No | ||
| 18 | ITPR2 | 2903 | 1.964 | 0.0349 | No | ||
| 19 | MAPK3 | 2925 | 1.958 | 0.0387 | No | ||
| 20 | BCAR1 | 2998 | 1.928 | 0.0396 | No | ||
| 21 | GRB2 | 3538 | 1.714 | 0.0148 | No | ||
| 22 | PTK2 | 4623 | 1.395 | -0.0403 | No | ||
| 23 | MAP4K1 | 4652 | 1.388 | -0.0384 | No | ||
| 24 | PIK3R1 | 4924 | 1.317 | -0.0497 | No | ||
| 25 | CDK7 | 5000 | 1.297 | -0.0505 | No | ||
| 26 | LYN | 5368 | 1.203 | -0.0673 | No | ||
| 27 | BLNK | 6505 | 0.938 | -0.1264 | No | ||
| 28 | CCNE1 | 6647 | 0.904 | -0.1317 | No | ||
| 29 | PTPN6 | 7405 | 0.740 | -0.1708 | No | ||
| 30 | NCK1 | 7424 | 0.736 | -0.1699 | No | ||
| 31 | HDAC5 | 7756 | 0.668 | -0.1861 | No | ||
| 32 | HCK | 7871 | 0.643 | -0.1907 | No | ||
| 33 | CD79A | 7962 | 0.622 | -0.1940 | No | ||
| 34 | CYCS | 8120 | 0.587 | -0.2010 | No | ||
| 35 | SOS1 | 8457 | 0.519 | -0.2179 | No | ||
| 36 | RAPGEF1 | 8488 | 0.511 | -0.2182 | No | ||
| 37 | GAB2 | 8545 | 0.496 | -0.2200 | No | ||
| 38 | AKT1 | 8696 | 0.467 | -0.2269 | No | ||
| 39 | PTPRC | 8852 | 0.434 | -0.2342 | No | ||
| 40 | CD19 | 8961 | 0.408 | -0.2390 | No | ||
| 41 | PLCG1 | 9454 | 0.300 | -0.2649 | No | ||
| 42 | REL | 9870 | 0.206 | -0.2868 | No | ||
| 43 | PRKCE | 10026 | 0.168 | -0.2947 | No | ||
| 44 | BTK | 10047 | 0.163 | -0.2954 | No | ||
| 45 | PTPN18 | 10256 | 0.113 | -0.3064 | No | ||
| 46 | CDK6 | 10258 | 0.112 | -0.3061 | No | ||
| 47 | SH3BP2 | 10414 | 0.067 | -0.3143 | No | ||
| 48 | PIK3AP1 | 11016 | -0.095 | -0.3466 | No | ||
| 49 | HCLS1 | 11238 | -0.157 | -0.3581 | No | ||
| 50 | CSK | 12109 | -0.409 | -0.4041 | No | ||
| 51 | PRKCD | 12195 | -0.435 | -0.4076 | No | ||
| 52 | NFATC2 | 12258 | -0.453 | -0.4098 | No | ||
| 53 | JUN | 12736 | -0.603 | -0.4341 | No | ||
| 54 | CR2 | 13751 | -0.990 | -0.4864 | No | ||
| 55 | MAP3K7 | 13920 | -1.056 | -0.4929 | No | ||
| 56 | MAPK4 | 13993 | -1.086 | -0.4940 | No | ||
| 57 | PRKD1 | 14257 | -1.214 | -0.5052 | No | ||
| 58 | CCNA2 | 14392 | -1.280 | -0.5092 | No | ||
| 59 | ACTR2 | 14626 | -1.388 | -0.5184 | No | ||
| 60 | ACTR3 | 14706 | -1.436 | -0.5190 | No | ||
| 61 | SYK | 14820 | -1.497 | -0.5214 | No | ||
| 62 | SOS2 | 14846 | -1.511 | -0.5189 | No | ||
| 63 | CASP9 | 14966 | -1.583 | -0.5214 | No | ||
| 64 | RASGRP3 | 15099 | -1.659 | -0.5244 | No | ||
| 65 | RPS6KB1 | 15174 | -1.716 | -0.5241 | No | ||
| 66 | PPP3CA | 15542 | -1.975 | -0.5390 | No | ||
| 67 | LCK | 15642 | -2.053 | -0.5392 | No | ||
| 68 | CBLB | 15678 | -2.083 | -0.5358 | No | ||
| 69 | PPP3R1 | 15684 | -2.086 | -0.5309 | No | ||
| 70 | VAV2 | 15777 | -2.167 | -0.5304 | No | ||
| 71 | IKBKB | 15915 | -2.297 | -0.5321 | No | ||
| 72 | CREB1 | 16167 | -2.549 | -0.5392 | No | ||
| 73 | CD81 | 16184 | -2.565 | -0.5337 | No | ||
| 74 | CHUK | 16242 | -2.624 | -0.5302 | No | ||
| 75 | MAPK8 | 16556 | -2.993 | -0.5396 | Yes | ||
| 76 | CCND3 | 16557 | -2.995 | -0.5321 | Yes | ||
| 77 | BCL2L11 | 16599 | -3.050 | -0.5266 | Yes | ||
| 78 | RASA1 | 16645 | -3.105 | -0.5213 | Yes | ||
| 79 | MAPK14 | 16698 | -3.190 | -0.5161 | Yes | ||
| 80 | RELA | 16711 | -3.206 | -0.5087 | Yes | ||
| 81 | NFKBIA | 16853 | -3.436 | -0.5077 | Yes | ||
| 82 | PTPN11 | 17014 | -3.663 | -0.5072 | Yes | ||
| 83 | MAPK1 | 17133 | -3.886 | -0.5038 | Yes | ||
| 84 | DAPP1 | 17159 | -3.922 | -0.4953 | Yes | ||
| 85 | CARD11 | 17214 | -4.022 | -0.4881 | Yes | ||
| 86 | IKBKG | 17233 | -4.051 | -0.4790 | Yes | ||
| 87 | WAS | 17259 | -4.085 | -0.4701 | Yes | ||
| 88 | FYN | 17414 | -4.454 | -0.4672 | Yes | ||
| 89 | ATF2 | 17460 | -4.560 | -0.4582 | Yes | ||
| 90 | BCL10 | 17583 | -4.820 | -0.4527 | Yes | ||
| 91 | GSK3B | 17591 | -4.835 | -0.4410 | Yes | ||
| 92 | CD5 | 17606 | -4.877 | -0.4295 | Yes | ||
| 93 | PRKCQ | 17644 | -4.969 | -0.4191 | Yes | ||
| 94 | CDK2 | 17662 | -5.028 | -0.4074 | Yes | ||
| 95 | RB1 | 17693 | -5.097 | -0.3962 | Yes | ||
| 96 | VAV1 | 17843 | -5.543 | -0.3904 | Yes | ||
| 97 | CD79B | 18022 | -6.195 | -0.3845 | Yes | ||
| 98 | PIP5K1C | 18088 | -6.520 | -0.3716 | Yes | ||
| 99 | CCND2 | 18106 | -6.603 | -0.3560 | Yes | ||
| 100 | CBL | 18121 | -6.671 | -0.3400 | Yes | ||
| 101 | LCP2 | 18154 | -6.845 | -0.3246 | Yes | ||
| 102 | GTF2I | 18179 | -6.977 | -0.3084 | Yes | ||
| 103 | PDK2 | 18205 | -7.150 | -0.2918 | Yes | ||
| 104 | CASP7 | 18227 | -7.255 | -0.2747 | Yes | ||
| 105 | NFATC1 | 18298 | -7.855 | -0.2588 | Yes | ||
| 106 | PTK2B | 18300 | -7.881 | -0.2391 | Yes | ||
| 107 | NFATC3 | 18304 | -7.953 | -0.2193 | Yes | ||
| 108 | PDPK1 | 18310 | -8.013 | -0.1995 | Yes | ||
| 109 | CRKL | 18345 | -8.316 | -0.1805 | Yes | ||
| 110 | ITK | 18503 | -10.655 | -0.1623 | Yes | ||
| 111 | BCL2 | 18517 | -11.183 | -0.1349 | Yes | ||
| 112 | NEDD9 | 18520 | -11.273 | -0.1068 | Yes | ||
| 113 | GAB1 | 18550 | -12.752 | -0.0764 | Yes | ||
| 114 | BCL6 | 18568 | -14.172 | -0.0417 | Yes | ||
| 115 | TEC | 18583 | -17.660 | 0.0018 | Yes |