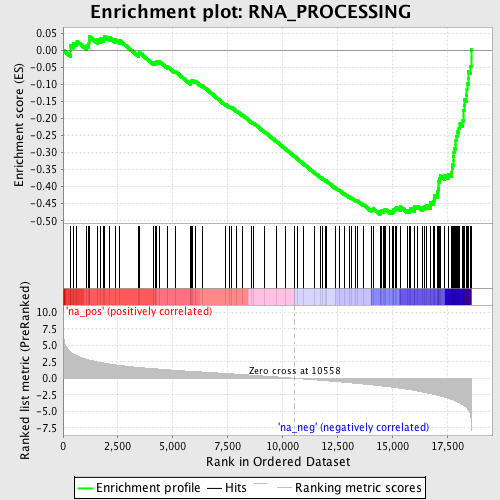
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | -0.48162362 |
| Normalized Enrichment Score (NES) | -2.1894844 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.006705207 |
| FWER p-Value | 0.018 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SF3A2 | 348 | 3.973 | -0.0029 | No | ||
| 2 | UPF1 | 349 | 3.967 | 0.0129 | No | ||
| 3 | GEMIN7 | 474 | 3.676 | 0.0209 | No | ||
| 4 | IGF2BP3 | 631 | 3.386 | 0.0260 | No | ||
| 5 | DDX54 | 1085 | 2.842 | 0.0128 | No | ||
| 6 | AARS | 1175 | 2.767 | 0.0191 | No | ||
| 7 | ZNF346 | 1180 | 2.763 | 0.0299 | No | ||
| 8 | LSM4 | 1193 | 2.755 | 0.0402 | No | ||
| 9 | RBPMS | 1557 | 2.484 | 0.0305 | No | ||
| 10 | METTL1 | 1692 | 2.395 | 0.0329 | No | ||
| 11 | SFRS8 | 1845 | 2.305 | 0.0338 | No | ||
| 12 | EXOSC3 | 1880 | 2.283 | 0.0411 | No | ||
| 13 | ELAVL4 | 2120 | 2.156 | 0.0368 | No | ||
| 14 | SNRPB | 2365 | 2.043 | 0.0318 | No | ||
| 15 | RBMY1A1 | 2587 | 1.955 | 0.0276 | No | ||
| 16 | SNRP70 | 3445 | 1.645 | -0.0121 | No | ||
| 17 | GEMIN5 | 3480 | 1.634 | -0.0074 | No | ||
| 18 | SF3A1 | 4128 | 1.452 | -0.0366 | No | ||
| 19 | NUFIP1 | 4215 | 1.433 | -0.0356 | No | ||
| 20 | ADAT1 | 4275 | 1.415 | -0.0331 | No | ||
| 21 | SNRPD2 | 4380 | 1.387 | -0.0332 | No | ||
| 22 | GEMIN4 | 4747 | 1.298 | -0.0478 | No | ||
| 23 | NOL3 | 5100 | 1.211 | -0.0620 | No | ||
| 24 | SFRS9 | 5800 | 1.053 | -0.0956 | No | ||
| 25 | DDX39 | 5825 | 1.049 | -0.0927 | No | ||
| 26 | U2AF1 | 5857 | 1.043 | -0.0902 | No | ||
| 27 | TXNL4A | 5890 | 1.037 | -0.0878 | No | ||
| 28 | DHX16 | 6037 | 1.008 | -0.0916 | No | ||
| 29 | FUSIP1 | 6344 | 0.942 | -0.1044 | No | ||
| 30 | NONO | 7418 | 0.724 | -0.1595 | No | ||
| 31 | RRP9 | 7597 | 0.686 | -0.1664 | No | ||
| 32 | EXOSC2 | 7661 | 0.673 | -0.1671 | No | ||
| 33 | LSM5 | 7921 | 0.618 | -0.1787 | No | ||
| 34 | SSB | 8189 | 0.559 | -0.1909 | No | ||
| 35 | PTBP1 | 8605 | 0.467 | -0.2115 | No | ||
| 36 | SRRM1 | 8679 | 0.449 | -0.2136 | No | ||
| 37 | SF3B4 | 9174 | 0.328 | -0.2390 | No | ||
| 38 | FARS2 | 9710 | 0.210 | -0.2671 | No | ||
| 39 | BCAS2 | 10137 | 0.110 | -0.2897 | No | ||
| 40 | SF3B3 | 10535 | 0.005 | -0.3111 | No | ||
| 41 | PPAN | 10558 | -0.000 | -0.3123 | No | ||
| 42 | DHX8 | 10693 | -0.034 | -0.3194 | No | ||
| 43 | CPSF3 | 10953 | -0.099 | -0.3330 | No | ||
| 44 | GEMIN6 | 11464 | -0.225 | -0.3597 | No | ||
| 45 | NUDT21 | 11717 | -0.291 | -0.3722 | No | ||
| 46 | SLBP | 11826 | -0.324 | -0.3767 | No | ||
| 47 | NOLA2 | 11958 | -0.356 | -0.3824 | No | ||
| 48 | SNRPA1 | 11988 | -0.365 | -0.3825 | No | ||
| 49 | BICD1 | 12419 | -0.481 | -0.4038 | No | ||
| 50 | SNRPN | 12577 | -0.526 | -0.4102 | No | ||
| 51 | SNRPB2 | 12828 | -0.598 | -0.4213 | No | ||
| 52 | RPS14 | 13033 | -0.651 | -0.4298 | No | ||
| 53 | HNRPF | 13122 | -0.681 | -0.4318 | No | ||
| 54 | SNRPF | 13326 | -0.748 | -0.4398 | No | ||
| 55 | FBL | 13403 | -0.771 | -0.4408 | No | ||
| 56 | LSM3 | 13671 | -0.854 | -0.4519 | No | ||
| 57 | RNPS1 | 14036 | -0.969 | -0.4677 | No | ||
| 58 | NCBP2 | 14072 | -0.983 | -0.4656 | No | ||
| 59 | SNRPD1 | 14126 | -1.004 | -0.4645 | No | ||
| 60 | SFRS7 | 14444 | -1.118 | -0.4772 | Yes | ||
| 61 | PRPF4B | 14461 | -1.123 | -0.4735 | Yes | ||
| 62 | NOLC1 | 14501 | -1.135 | -0.4711 | Yes | ||
| 63 | PPARGC1A | 14606 | -1.174 | -0.4721 | Yes | ||
| 64 | CSTF1 | 14626 | -1.183 | -0.4684 | Yes | ||
| 65 | EFTUD2 | 14682 | -1.207 | -0.4665 | Yes | ||
| 66 | EXOSC7 | 14886 | -1.283 | -0.4724 | Yes | ||
| 67 | PHF5A | 15018 | -1.345 | -0.4741 | Yes | ||
| 68 | RNMT | 15059 | -1.364 | -0.4708 | Yes | ||
| 69 | CRNKL1 | 15070 | -1.368 | -0.4659 | Yes | ||
| 70 | SIP1 | 15136 | -1.395 | -0.4638 | Yes | ||
| 71 | HNRPD | 15203 | -1.430 | -0.4617 | Yes | ||
| 72 | CPSF1 | 15369 | -1.506 | -0.4646 | Yes | ||
| 73 | ZNF638 | 15380 | -1.511 | -0.4591 | Yes | ||
| 74 | PRPF8 | 15690 | -1.651 | -0.4692 | Yes | ||
| 75 | RBM3 | 15809 | -1.712 | -0.4687 | Yes | ||
| 76 | ASCC3L1 | 15842 | -1.729 | -0.4636 | Yes | ||
| 77 | SNRPG | 16002 | -1.836 | -0.4648 | Yes | ||
| 78 | SNW1 | 16009 | -1.840 | -0.4578 | Yes | ||
| 79 | RBM6 | 16137 | -1.907 | -0.4570 | Yes | ||
| 80 | SNRPD3 | 16367 | -2.054 | -0.4612 | Yes | ||
| 81 | SF3B2 | 16465 | -2.115 | -0.4580 | Yes | ||
| 82 | PABPC4 | 16565 | -2.188 | -0.4546 | Yes | ||
| 83 | INTS6 | 16735 | -2.298 | -0.4546 | Yes | ||
| 84 | SYNCRIP | 16752 | -2.313 | -0.4462 | Yes | ||
| 85 | HNRPUL1 | 16880 | -2.408 | -0.4435 | Yes | ||
| 86 | RNGTT | 16910 | -2.429 | -0.4353 | Yes | ||
| 87 | CSTF3 | 16935 | -2.449 | -0.4269 | Yes | ||
| 88 | DHX38 | 17055 | -2.546 | -0.4231 | Yes | ||
| 89 | TRNT1 | 17072 | -2.560 | -0.4138 | Yes | ||
| 90 | KIN | 17095 | -2.581 | -0.4046 | Yes | ||
| 91 | PRPF31 | 17116 | -2.593 | -0.3954 | Yes | ||
| 92 | RBMS1 | 17117 | -2.594 | -0.3850 | Yes | ||
| 93 | SFRS10 | 17151 | -2.636 | -0.3763 | Yes | ||
| 94 | SRPK1 | 17180 | -2.660 | -0.3672 | Yes | ||
| 95 | PABPC1 | 17401 | -2.865 | -0.3676 | Yes | ||
| 96 | CSTF2 | 17550 | -3.014 | -0.3636 | Yes | ||
| 97 | RBM5 | 17702 | -3.169 | -0.3591 | Yes | ||
| 98 | NSUN2 | 17724 | -3.194 | -0.3475 | Yes | ||
| 99 | SFRS5 | 17737 | -3.212 | -0.3353 | Yes | ||
| 100 | SFRS2 | 17780 | -3.269 | -0.3245 | Yes | ||
| 101 | NOL5A | 17783 | -3.271 | -0.3115 | Yes | ||
| 102 | SF3A3 | 17808 | -3.301 | -0.2996 | Yes | ||
| 103 | SFRS2IP | 17828 | -3.326 | -0.2874 | Yes | ||
| 104 | CDC2L2 | 17899 | -3.436 | -0.2774 | Yes | ||
| 105 | GRSF1 | 17902 | -3.442 | -0.2638 | Yes | ||
| 106 | DGCR8 | 17938 | -3.501 | -0.2517 | Yes | ||
| 107 | KHDRBS1 | 17970 | -3.552 | -0.2392 | Yes | ||
| 108 | SFPQ | 18040 | -3.653 | -0.2283 | Yes | ||
| 109 | SPOP | 18087 | -3.732 | -0.2159 | Yes | ||
| 110 | DHX15 | 18222 | -3.982 | -0.2073 | Yes | ||
| 111 | SFRS6 | 18258 | -4.074 | -0.1929 | Yes | ||
| 112 | HNRPDL | 18259 | -4.077 | -0.1766 | Yes | ||
| 113 | SRPK2 | 18276 | -4.121 | -0.1610 | Yes | ||
| 114 | CUGBP1 | 18289 | -4.148 | -0.1451 | Yes | ||
| 115 | POP4 | 18381 | -4.402 | -0.1324 | Yes | ||
| 116 | SFRS1 | 18407 | -4.486 | -0.1159 | Yes | ||
| 117 | USP39 | 18409 | -4.505 | -0.0979 | Yes | ||
| 118 | DUSP11 | 18459 | -4.673 | -0.0819 | Yes | ||
| 119 | IVNS1ABP | 18464 | -4.681 | -0.0634 | Yes | ||
| 120 | PRPF3 | 18575 | -5.684 | -0.0467 | Yes | ||
| 121 | DBR1 | 18598 | -6.102 | -0.0235 | Yes | ||
| 122 | DDX20 | 18599 | -6.114 | 0.0009 | Yes |
