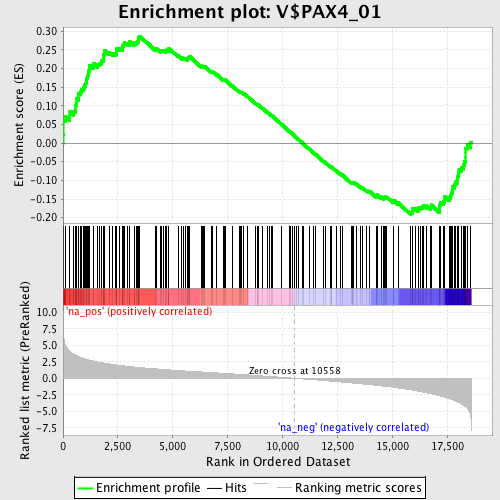
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$PAX4_01 |
| Enrichment Score (ES) | 0.28670058 |
| Normalized Enrichment Score (NES) | 1.37598 |
| Nominal p-value | 0.013618677 |
| FDR q-value | 0.61469674 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DTX1 | 2 | 8.089 | 0.0235 | Yes | ||
| 2 | HES1 | 19 | 6.611 | 0.0419 | Yes | ||
| 3 | RNF10 | 20 | 6.596 | 0.0611 | Yes | ||
| 4 | HTF9C | 101 | 5.047 | 0.0715 | Yes | ||
| 5 | CAPN5 | 295 | 4.145 | 0.0731 | Yes | ||
| 6 | FBXL19 | 304 | 4.109 | 0.0846 | Yes | ||
| 7 | OAZ2 | 482 | 3.654 | 0.0857 | Yes | ||
| 8 | SUOX | 567 | 3.501 | 0.0913 | Yes | ||
| 9 | PLAGL1 | 570 | 3.494 | 0.1014 | Yes | ||
| 10 | NCDN | 603 | 3.437 | 0.1097 | Yes | ||
| 11 | DGKZ | 620 | 3.400 | 0.1187 | Yes | ||
| 12 | HOXB1 | 688 | 3.297 | 0.1247 | Yes | ||
| 13 | RPS6KA4 | 694 | 3.288 | 0.1340 | Yes | ||
| 14 | ETV5 | 791 | 3.153 | 0.1380 | Yes | ||
| 15 | LBX1 | 832 | 3.108 | 0.1449 | Yes | ||
| 16 | PTPN23 | 921 | 3.018 | 0.1489 | Yes | ||
| 17 | RING1 | 956 | 2.983 | 0.1558 | Yes | ||
| 18 | EIF5A | 1032 | 2.891 | 0.1601 | Yes | ||
| 19 | CTDSP1 | 1064 | 2.863 | 0.1668 | Yes | ||
| 20 | AAMP | 1078 | 2.846 | 0.1744 | Yes | ||
| 21 | TITF1 | 1092 | 2.839 | 0.1819 | Yes | ||
| 22 | NEUROG1 | 1138 | 2.802 | 0.1877 | Yes | ||
| 23 | LRP2 | 1160 | 2.779 | 0.1946 | Yes | ||
| 24 | PRKAG1 | 1192 | 2.756 | 0.2010 | Yes | ||
| 25 | BAT3 | 1212 | 2.739 | 0.2079 | Yes | ||
| 26 | PIK3CD | 1377 | 2.615 | 0.2067 | Yes | ||
| 27 | TRERF1 | 1394 | 2.602 | 0.2134 | Yes | ||
| 28 | FBXO36 | 1585 | 2.468 | 0.2103 | Yes | ||
| 29 | POFUT1 | 1647 | 2.428 | 0.2140 | Yes | ||
| 30 | POU3F3 | 1729 | 2.372 | 0.2165 | Yes | ||
| 31 | HOXA10 | 1762 | 2.351 | 0.2217 | Yes | ||
| 32 | SART3 | 1836 | 2.315 | 0.2245 | Yes | ||
| 33 | PTPRG | 1851 | 2.301 | 0.2304 | Yes | ||
| 34 | MNT | 1854 | 2.299 | 0.2370 | Yes | ||
| 35 | ASB7 | 1895 | 2.275 | 0.2415 | Yes | ||
| 36 | ANGPT1 | 1903 | 2.268 | 0.2477 | Yes | ||
| 37 | CNTFR | 2102 | 2.166 | 0.2433 | Yes | ||
| 38 | STX1B2 | 2267 | 2.090 | 0.2405 | Yes | ||
| 39 | RRAS | 2375 | 2.041 | 0.2406 | Yes | ||
| 40 | CHKB | 2427 | 2.022 | 0.2437 | Yes | ||
| 41 | LEMD2 | 2428 | 2.022 | 0.2496 | Yes | ||
| 42 | PGF | 2451 | 2.011 | 0.2543 | Yes | ||
| 43 | DAAM2 | 2586 | 1.955 | 0.2527 | Yes | ||
| 44 | CIZ1 | 2692 | 1.906 | 0.2526 | Yes | ||
| 45 | PHKB | 2709 | 1.900 | 0.2572 | Yes | ||
| 46 | TPM1 | 2722 | 1.894 | 0.2621 | Yes | ||
| 47 | SALL1 | 2767 | 1.878 | 0.2652 | Yes | ||
| 48 | CIC | 2775 | 1.875 | 0.2703 | Yes | ||
| 49 | PER2 | 2915 | 1.815 | 0.2680 | Yes | ||
| 50 | ABHD2 | 3021 | 1.773 | 0.2675 | Yes | ||
| 51 | SORCS3 | 3031 | 1.771 | 0.2722 | Yes | ||
| 52 | AP1B1 | 3234 | 1.710 | 0.2662 | Yes | ||
| 53 | EFHD1 | 3269 | 1.700 | 0.2693 | Yes | ||
| 54 | ESRRA | 3348 | 1.673 | 0.2700 | Yes | ||
| 55 | PAQR4 | 3397 | 1.661 | 0.2722 | Yes | ||
| 56 | HOXB6 | 3424 | 1.651 | 0.2756 | Yes | ||
| 57 | RKHD3 | 3438 | 1.647 | 0.2797 | Yes | ||
| 58 | NTRK3 | 3447 | 1.644 | 0.2841 | Yes | ||
| 59 | KLC2 | 3487 | 1.631 | 0.2867 | Yes | ||
| 60 | BRUNOL6 | 4195 | 1.437 | 0.2525 | No | ||
| 61 | CDK9 | 4233 | 1.427 | 0.2547 | No | ||
| 62 | SLC35B1 | 4448 | 1.369 | 0.2471 | No | ||
| 63 | HOXA1 | 4500 | 1.357 | 0.2483 | No | ||
| 64 | DNM1 | 4573 | 1.338 | 0.2482 | No | ||
| 65 | BDNF | 4682 | 1.313 | 0.2462 | No | ||
| 66 | BCL11A | 4692 | 1.312 | 0.2496 | No | ||
| 67 | ARF6 | 4786 | 1.291 | 0.2483 | No | ||
| 68 | CDX4 | 4803 | 1.286 | 0.2511 | No | ||
| 69 | RAB33A | 4818 | 1.282 | 0.2541 | No | ||
| 70 | GPR37L1 | 5243 | 1.174 | 0.2345 | No | ||
| 71 | SEMA3B | 5416 | 1.135 | 0.2285 | No | ||
| 72 | FGD1 | 5502 | 1.115 | 0.2272 | No | ||
| 73 | NR0B2 | 5576 | 1.100 | 0.2264 | No | ||
| 74 | ARHGEF12 | 5650 | 1.086 | 0.2256 | No | ||
| 75 | PCTK1 | 5667 | 1.084 | 0.2279 | No | ||
| 76 | SLC38A3 | 5707 | 1.073 | 0.2289 | No | ||
| 77 | CACNA1D | 5747 | 1.064 | 0.2299 | No | ||
| 78 | GNB2 | 5761 | 1.062 | 0.2323 | No | ||
| 79 | DLL4 | 6323 | 0.946 | 0.2046 | No | ||
| 80 | TNXB | 6337 | 0.944 | 0.2067 | No | ||
| 81 | NRXN1 | 6408 | 0.928 | 0.2056 | No | ||
| 82 | C1QL1 | 6454 | 0.919 | 0.2058 | No | ||
| 83 | PHF15 | 6741 | 0.863 | 0.1928 | No | ||
| 84 | MXD4 | 6811 | 0.849 | 0.1915 | No | ||
| 85 | HNRPR | 7002 | 0.811 | 0.1836 | No | ||
| 86 | DDIT3 | 7312 | 0.746 | 0.1690 | No | ||
| 87 | ACOX2 | 7344 | 0.740 | 0.1695 | No | ||
| 88 | GRIN2A | 7385 | 0.732 | 0.1695 | No | ||
| 89 | TRIM3 | 7704 | 0.666 | 0.1541 | No | ||
| 90 | PDGFB | 8041 | 0.591 | 0.1376 | No | ||
| 91 | SLC16A10 | 8087 | 0.582 | 0.1369 | No | ||
| 92 | CTF1 | 8123 | 0.574 | 0.1367 | No | ||
| 93 | IRX2 | 8239 | 0.547 | 0.1320 | No | ||
| 94 | ZHX2 | 8396 | 0.512 | 0.1251 | No | ||
| 95 | CDH16 | 8753 | 0.431 | 0.1070 | No | ||
| 96 | EFNA5 | 8849 | 0.407 | 0.1030 | No | ||
| 97 | JARID2 | 8859 | 0.404 | 0.1037 | No | ||
| 98 | CCNI | 8905 | 0.393 | 0.1024 | No | ||
| 99 | GRIK1 | 9086 | 0.350 | 0.0937 | No | ||
| 100 | GALNT10 | 9302 | 0.303 | 0.0829 | No | ||
| 101 | PRDM1 | 9408 | 0.281 | 0.0780 | No | ||
| 102 | HOXD10 | 9500 | 0.262 | 0.0738 | No | ||
| 103 | KCNK13 | 9544 | 0.251 | 0.0722 | No | ||
| 104 | RBP4 | 9974 | 0.147 | 0.0494 | No | ||
| 105 | HMGN2 | 10306 | 0.061 | 0.0316 | No | ||
| 106 | ZIC2 | 10343 | 0.053 | 0.0298 | No | ||
| 107 | PACS1 | 10433 | 0.029 | 0.0251 | No | ||
| 108 | SP4 | 10550 | 0.002 | 0.0188 | No | ||
| 109 | CPT1B | 10653 | -0.022 | 0.0133 | No | ||
| 110 | SLC25A28 | 10708 | -0.038 | 0.0105 | No | ||
| 111 | SLC4A1 | 10904 | -0.087 | 0.0002 | No | ||
| 112 | DNAJB5 | 10940 | -0.096 | -0.0014 | No | ||
| 113 | SMAD1 | 11244 | -0.174 | -0.0174 | No | ||
| 114 | XPO1 | 11398 | -0.210 | -0.0251 | No | ||
| 115 | LRP1 | 11519 | -0.238 | -0.0309 | No | ||
| 116 | PBX1 | 11859 | -0.333 | -0.0483 | No | ||
| 117 | SPRY2 | 11953 | -0.354 | -0.0523 | No | ||
| 118 | MMP11 | 12183 | -0.417 | -0.0635 | No | ||
| 119 | LIN28 | 12211 | -0.424 | -0.0637 | No | ||
| 120 | CDX1 | 12236 | -0.430 | -0.0638 | No | ||
| 121 | PHF12 | 12465 | -0.493 | -0.0747 | No | ||
| 122 | HOXA7 | 12623 | -0.539 | -0.0817 | No | ||
| 123 | SSH2 | 12718 | -0.569 | -0.0851 | No | ||
| 124 | RBMS3 | 13141 | -0.686 | -0.1060 | No | ||
| 125 | PCF11 | 13200 | -0.708 | -0.1071 | No | ||
| 126 | COL27A1 | 13201 | -0.708 | -0.1050 | No | ||
| 127 | EVX1 | 13239 | -0.718 | -0.1049 | No | ||
| 128 | NFKBIA | 13366 | -0.761 | -0.1095 | No | ||
| 129 | GTF2IRD1 | 13550 | -0.814 | -0.1171 | No | ||
| 130 | ATOH1 | 13639 | -0.842 | -0.1194 | No | ||
| 131 | AMFR | 13831 | -0.901 | -0.1272 | No | ||
| 132 | PLAGL2 | 13943 | -0.940 | -0.1304 | No | ||
| 133 | IL17B | 13981 | -0.951 | -0.1297 | No | ||
| 134 | APBA2BP | 14265 | -1.049 | -0.1420 | No | ||
| 135 | ZFYVE27 | 14282 | -1.058 | -0.1397 | No | ||
| 136 | NR4A3 | 14316 | -1.070 | -0.1384 | No | ||
| 137 | CPNE1 | 14488 | -1.129 | -0.1444 | No | ||
| 138 | NRD1 | 14604 | -1.174 | -0.1472 | No | ||
| 139 | ISCU | 14631 | -1.185 | -0.1452 | No | ||
| 140 | NF2 | 14671 | -1.202 | -0.1438 | No | ||
| 141 | ACSL3 | 14755 | -1.230 | -0.1447 | No | ||
| 142 | PDE4D | 15041 | -1.357 | -0.1562 | No | ||
| 143 | H1FX | 15079 | -1.371 | -0.1542 | No | ||
| 144 | E2F1 | 15267 | -1.457 | -0.1601 | No | ||
| 145 | DNAJC11 | 15846 | -1.732 | -0.1864 | No | ||
| 146 | UBTF | 15914 | -1.772 | -0.1849 | No | ||
| 147 | NFATC4 | 15924 | -1.780 | -0.1802 | No | ||
| 148 | PSIP1 | 15927 | -1.781 | -0.1751 | No | ||
| 149 | ANKMY2 | 16041 | -1.852 | -0.1758 | No | ||
| 150 | ADAMTSL1 | 16184 | -1.934 | -0.1779 | No | ||
| 151 | PICALM | 16198 | -1.942 | -0.1730 | No | ||
| 152 | GADD45G | 16295 | -1.999 | -0.1723 | No | ||
| 153 | WNT10A | 16371 | -2.056 | -0.1704 | No | ||
| 154 | RBBP6 | 16408 | -2.077 | -0.1663 | No | ||
| 155 | PABPC4 | 16565 | -2.188 | -0.1684 | No | ||
| 156 | SYNCRIP | 16752 | -2.313 | -0.1717 | No | ||
| 157 | CTBP2 | 16769 | -2.322 | -0.1658 | No | ||
| 158 | TAF11 | 17144 | -2.628 | -0.1785 | No | ||
| 159 | EIF2C2 | 17153 | -2.637 | -0.1712 | No | ||
| 160 | HIF3A | 17171 | -2.652 | -0.1644 | No | ||
| 161 | MAPK10 | 17207 | -2.691 | -0.1585 | No | ||
| 162 | YWHAZ | 17324 | -2.796 | -0.1566 | No | ||
| 163 | ADIPOR2 | 17380 | -2.838 | -0.1513 | No | ||
| 164 | PABPC1 | 17401 | -2.865 | -0.1441 | No | ||
| 165 | MAPT | 17609 | -3.078 | -0.1463 | No | ||
| 166 | APPBP1 | 17649 | -3.110 | -0.1394 | No | ||
| 167 | ZCCHC9 | 17684 | -3.150 | -0.1320 | No | ||
| 168 | KLF13 | 17731 | -3.203 | -0.1252 | No | ||
| 169 | GTF3C2 | 17766 | -3.248 | -0.1176 | No | ||
| 170 | MAPRE1 | 17831 | -3.327 | -0.1113 | No | ||
| 171 | EIF4A1 | 17880 | -3.409 | -0.1040 | No | ||
| 172 | RANBP1 | 17962 | -3.537 | -0.0981 | No | ||
| 173 | TPR | 17981 | -3.575 | -0.0887 | No | ||
| 174 | MAML1 | 18015 | -3.619 | -0.0799 | No | ||
| 175 | ZDHHC5 | 18042 | -3.656 | -0.0706 | No | ||
| 176 | TNRC15 | 18145 | -3.835 | -0.0650 | No | ||
| 177 | KPNB1 | 18242 | -4.033 | -0.0585 | No | ||
| 178 | ELAVL1 | 18282 | -4.139 | -0.0485 | No | ||
| 179 | TAF5 | 18333 | -4.253 | -0.0388 | No | ||
| 180 | MXI1 | 18336 | -4.258 | -0.0265 | No | ||
| 181 | EED | 18339 | -4.265 | -0.0142 | No | ||
| 182 | USP39 | 18409 | -4.505 | -0.0048 | No | ||
| 183 | SLC30A5 | 18566 | -5.483 | 0.0027 | No |
