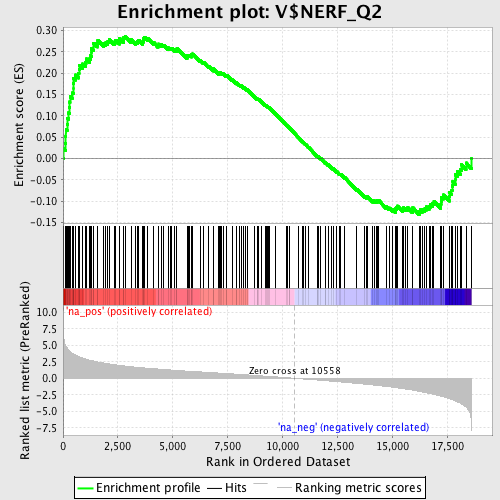
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$NERF_Q2 |
| Enrichment Score (ES) | 0.28661767 |
| Normalized Enrichment Score (NES) | 1.3641249 |
| Nominal p-value | 0.016513761 |
| FDR q-value | 0.62248117 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RGS3 | 12 | 7.042 | 0.0234 | Yes | ||
| 2 | HTF9C | 101 | 5.047 | 0.0359 | Yes | ||
| 3 | POLD4 | 122 | 4.910 | 0.0517 | Yes | ||
| 4 | RASAL1 | 131 | 4.856 | 0.0678 | Yes | ||
| 5 | HMGA1 | 184 | 4.595 | 0.0807 | Yes | ||
| 6 | LANCL1 | 211 | 4.440 | 0.0945 | Yes | ||
| 7 | MAP4K1 | 254 | 4.300 | 0.1069 | Yes | ||
| 8 | DOK1 | 284 | 4.174 | 0.1197 | Yes | ||
| 9 | AGPAT1 | 306 | 4.100 | 0.1325 | Yes | ||
| 10 | ROM1 | 314 | 4.073 | 0.1461 | Yes | ||
| 11 | SH2D2A | 413 | 3.776 | 0.1537 | Yes | ||
| 12 | CORO1A | 460 | 3.694 | 0.1638 | Yes | ||
| 13 | USP5 | 463 | 3.687 | 0.1764 | Yes | ||
| 14 | FGR | 477 | 3.667 | 0.1882 | Yes | ||
| 15 | MAPK3 | 561 | 3.505 | 0.1957 | Yes | ||
| 16 | RPS6KA4 | 694 | 3.288 | 0.1998 | Yes | ||
| 17 | PTPN6 | 726 | 3.242 | 0.2092 | Yes | ||
| 18 | STAT5A | 754 | 3.208 | 0.2187 | Yes | ||
| 19 | ARRB2 | 896 | 3.043 | 0.2215 | Yes | ||
| 20 | DEF6 | 1027 | 2.895 | 0.2243 | Yes | ||
| 21 | TRAPPC3 | 1045 | 2.883 | 0.2333 | Yes | ||
| 22 | ITPR3 | 1218 | 2.734 | 0.2333 | Yes | ||
| 23 | NRXN3 | 1252 | 2.712 | 0.2408 | Yes | ||
| 24 | CREB3 | 1281 | 2.693 | 0.2485 | Yes | ||
| 25 | TLN1 | 1286 | 2.687 | 0.2575 | Yes | ||
| 26 | TWIST1 | 1391 | 2.604 | 0.2607 | Yes | ||
| 27 | C1QTNF6 | 1392 | 2.604 | 0.2696 | Yes | ||
| 28 | ARHGDIB | 1550 | 2.488 | 0.2696 | Yes | ||
| 29 | FBXO36 | 1585 | 2.468 | 0.2762 | Yes | ||
| 30 | EGR3 | 1848 | 2.302 | 0.2699 | Yes | ||
| 31 | PPP1R9B | 1951 | 2.244 | 0.2721 | Yes | ||
| 32 | SPRED2 | 2040 | 2.203 | 0.2748 | Yes | ||
| 33 | MDFI | 2109 | 2.161 | 0.2785 | Yes | ||
| 34 | PEX16 | 2343 | 2.053 | 0.2729 | Yes | ||
| 35 | FBS1 | 2392 | 2.033 | 0.2773 | Yes | ||
| 36 | CNTNAP1 | 2564 | 1.964 | 0.2747 | Yes | ||
| 37 | JUNB | 2580 | 1.958 | 0.2806 | Yes | ||
| 38 | CENTD2 | 2748 | 1.887 | 0.2780 | Yes | ||
| 39 | SALL1 | 2767 | 1.878 | 0.2835 | Yes | ||
| 40 | TENC1 | 2827 | 1.851 | 0.2866 | Yes | ||
| 41 | TGIF2 | 3096 | 1.750 | 0.2781 | No | ||
| 42 | GGTL3 | 3310 | 1.688 | 0.2723 | No | ||
| 43 | DPP3 | 3384 | 1.664 | 0.2740 | No | ||
| 44 | NTRK3 | 3447 | 1.644 | 0.2763 | No | ||
| 45 | LYPLA2 | 3625 | 1.589 | 0.2722 | No | ||
| 46 | MMRN2 | 3646 | 1.583 | 0.2765 | No | ||
| 47 | LSAMP | 3675 | 1.576 | 0.2804 | No | ||
| 48 | FURIN | 3707 | 1.566 | 0.2840 | No | ||
| 49 | STAT5B | 3867 | 1.521 | 0.2806 | No | ||
| 50 | CLIC1 | 4122 | 1.453 | 0.2718 | No | ||
| 51 | ITPKB | 4325 | 1.403 | 0.2657 | No | ||
| 52 | FGFR2 | 4367 | 1.390 | 0.2682 | No | ||
| 53 | LST1 | 4475 | 1.362 | 0.2671 | No | ||
| 54 | NFIA | 4585 | 1.334 | 0.2657 | No | ||
| 55 | CD79B | 4790 | 1.290 | 0.2591 | No | ||
| 56 | RPS3 | 4877 | 1.264 | 0.2588 | No | ||
| 57 | ADAMTS4 | 4959 | 1.245 | 0.2586 | No | ||
| 58 | PITX2 | 5098 | 1.211 | 0.2553 | No | ||
| 59 | FXYD5 | 5179 | 1.192 | 0.2550 | No | ||
| 60 | GRM3 | 5189 | 1.190 | 0.2586 | No | ||
| 61 | TMEM4 | 5649 | 1.086 | 0.2374 | No | ||
| 62 | POU6F2 | 5654 | 1.086 | 0.2409 | No | ||
| 63 | UQCRH | 5713 | 1.072 | 0.2415 | No | ||
| 64 | BAZ2A | 5782 | 1.058 | 0.2414 | No | ||
| 65 | FLT1 | 5852 | 1.044 | 0.2412 | No | ||
| 66 | FCER1G | 5859 | 1.043 | 0.2445 | No | ||
| 67 | ROBO4 | 5917 | 1.033 | 0.2449 | No | ||
| 68 | ARHGAP8 | 6263 | 0.960 | 0.2295 | No | ||
| 69 | PAX6 | 6396 | 0.930 | 0.2255 | No | ||
| 70 | SEMA4C | 6636 | 0.883 | 0.2156 | No | ||
| 71 | SH3TC1 | 6835 | 0.843 | 0.2078 | No | ||
| 72 | MID1 | 6841 | 0.841 | 0.2104 | No | ||
| 73 | NPY | 7084 | 0.793 | 0.2000 | No | ||
| 74 | ZFPM1 | 7117 | 0.786 | 0.2009 | No | ||
| 75 | RHOV | 7151 | 0.781 | 0.2018 | No | ||
| 76 | CNNM1 | 7229 | 0.766 | 0.2002 | No | ||
| 77 | DDIT3 | 7312 | 0.746 | 0.1984 | No | ||
| 78 | MPL | 7438 | 0.721 | 0.1940 | No | ||
| 79 | HOXC4 | 7463 | 0.714 | 0.1952 | No | ||
| 80 | FOSL1 | 7706 | 0.666 | 0.1843 | No | ||
| 81 | BMP4 | 7888 | 0.625 | 0.1767 | No | ||
| 82 | CPNE8 | 7892 | 0.623 | 0.1786 | No | ||
| 83 | ST7L | 8052 | 0.589 | 0.1720 | No | ||
| 84 | CAP1 | 8119 | 0.574 | 0.1704 | No | ||
| 85 | NDUFS2 | 8236 | 0.549 | 0.1660 | No | ||
| 86 | TRO | 8309 | 0.532 | 0.1639 | No | ||
| 87 | ZHX2 | 8396 | 0.512 | 0.1610 | No | ||
| 88 | POU4F3 | 8734 | 0.434 | 0.1442 | No | ||
| 89 | CDK5 | 8854 | 0.405 | 0.1392 | No | ||
| 90 | LAT | 8871 | 0.400 | 0.1397 | No | ||
| 91 | HCLS1 | 8909 | 0.392 | 0.1390 | No | ||
| 92 | SLC4A2 | 9042 | 0.359 | 0.1331 | No | ||
| 93 | CD79A | 9216 | 0.319 | 0.1248 | No | ||
| 94 | TUSC3 | 9249 | 0.312 | 0.1241 | No | ||
| 95 | LYN | 9298 | 0.303 | 0.1226 | No | ||
| 96 | JAG1 | 9373 | 0.288 | 0.1196 | No | ||
| 97 | SNCG | 9428 | 0.277 | 0.1176 | No | ||
| 98 | HIC1 | 9683 | 0.216 | 0.1045 | No | ||
| 99 | KCND1 | 10195 | 0.092 | 0.0772 | No | ||
| 100 | OLFML2A | 10231 | 0.083 | 0.0756 | No | ||
| 101 | HMGN2 | 10306 | 0.061 | 0.0718 | No | ||
| 102 | SHC3 | 10329 | 0.056 | 0.0708 | No | ||
| 103 | PIGW | 10709 | -0.038 | 0.0503 | No | ||
| 104 | SLCO2B1 | 10898 | -0.086 | 0.0404 | No | ||
| 105 | EFNA1 | 10939 | -0.096 | 0.0386 | No | ||
| 106 | HYAL2 | 11042 | -0.123 | 0.0335 | No | ||
| 107 | PTPRN | 11188 | -0.160 | 0.0262 | No | ||
| 108 | LYL1 | 11598 | -0.260 | 0.0049 | No | ||
| 109 | ATOH8 | 11602 | -0.260 | 0.0056 | No | ||
| 110 | SLITRK6 | 11654 | -0.276 | 0.0038 | No | ||
| 111 | HOXB9 | 11746 | -0.299 | -0.0001 | No | ||
| 112 | PLCB2 | 11943 | -0.352 | -0.0095 | No | ||
| 113 | PEX11B | 11950 | -0.354 | -0.0086 | No | ||
| 114 | PIK3CG | 12083 | -0.391 | -0.0144 | No | ||
| 115 | CAPZA1 | 12245 | -0.433 | -0.0217 | No | ||
| 116 | UBE2L3 | 12302 | -0.447 | -0.0232 | No | ||
| 117 | SDCCAG8 | 12480 | -0.497 | -0.0311 | No | ||
| 118 | SRPR | 12607 | -0.533 | -0.0361 | No | ||
| 119 | PAK3 | 12650 | -0.547 | -0.0365 | No | ||
| 120 | GMFB | 12835 | -0.599 | -0.0444 | No | ||
| 121 | DIABLO | 13387 | -0.766 | -0.0717 | No | ||
| 122 | GPR132 | 13741 | -0.876 | -0.0878 | No | ||
| 123 | CHD2 | 13845 | -0.907 | -0.0903 | No | ||
| 124 | SLC30A7 | 13859 | -0.912 | -0.0879 | No | ||
| 125 | STARD8 | 14116 | -1.000 | -0.0983 | No | ||
| 126 | IKBKB | 14196 | -1.028 | -0.0991 | No | ||
| 127 | FKBP10 | 14264 | -1.049 | -0.0991 | No | ||
| 128 | ERG | 14317 | -1.070 | -0.0983 | No | ||
| 129 | ZNF23 | 14372 | -1.091 | -0.0975 | No | ||
| 130 | ATP5H | 14729 | -1.220 | -0.1126 | No | ||
| 131 | TPP2 | 14866 | -1.276 | -0.1156 | No | ||
| 132 | ATP1A1 | 15022 | -1.348 | -0.1194 | No | ||
| 133 | LOXL3 | 15159 | -1.405 | -0.1220 | No | ||
| 134 | ACTR3 | 15164 | -1.406 | -0.1174 | No | ||
| 135 | CDCA3 | 15207 | -1.433 | -0.1147 | No | ||
| 136 | RBBP7 | 15228 | -1.441 | -0.1109 | No | ||
| 137 | MAP2K6 | 15459 | -1.542 | -0.1181 | No | ||
| 138 | TFRC | 15504 | -1.563 | -0.1151 | No | ||
| 139 | GOLGA7 | 15620 | -1.616 | -0.1158 | No | ||
| 140 | ELK3 | 15714 | -1.664 | -0.1152 | No | ||
| 141 | UBTF | 15914 | -1.772 | -0.1199 | No | ||
| 142 | MCTS1 | 15943 | -1.792 | -0.1153 | No | ||
| 143 | RASA1 | 16230 | -1.958 | -0.1241 | No | ||
| 144 | PCDH7 | 16278 | -1.986 | -0.1198 | No | ||
| 145 | EBAG9 | 16377 | -2.059 | -0.1181 | No | ||
| 146 | CAMK1D | 16492 | -2.132 | -0.1170 | No | ||
| 147 | ELOVL5 | 16556 | -2.181 | -0.1130 | No | ||
| 148 | NLK | 16679 | -2.261 | -0.1118 | No | ||
| 149 | VCL | 16727 | -2.291 | -0.1065 | No | ||
| 150 | TCF12 | 16830 | -2.371 | -0.1040 | No | ||
| 151 | STARD13 | 16903 | -2.426 | -0.0996 | No | ||
| 152 | HN1 | 17215 | -2.697 | -0.1072 | No | ||
| 153 | SECISBP2 | 17225 | -2.704 | -0.0984 | No | ||
| 154 | DPP8 | 17249 | -2.727 | -0.0903 | No | ||
| 155 | BTBD12 | 17329 | -2.800 | -0.0850 | No | ||
| 156 | CALM2 | 17594 | -3.060 | -0.0889 | No | ||
| 157 | FCGR2B | 17613 | -3.081 | -0.0793 | No | ||
| 158 | NFATC3 | 17715 | -3.185 | -0.0739 | No | ||
| 159 | KLF13 | 17731 | -3.203 | -0.0638 | No | ||
| 160 | EXTL2 | 17764 | -3.247 | -0.0544 | No | ||
| 161 | MYOHD1 | 17869 | -3.394 | -0.0484 | No | ||
| 162 | TOPORS | 17875 | -3.404 | -0.0370 | No | ||
| 163 | RANBP1 | 17962 | -3.537 | -0.0296 | No | ||
| 164 | LRRFIP2 | 18121 | -3.788 | -0.0252 | No | ||
| 165 | UCHL5 | 18165 | -3.866 | -0.0143 | No | ||
| 166 | BCOR | 18364 | -4.333 | -0.0102 | No | ||
| 167 | FREQ | 18608 | -6.962 | 0.0004 | No |
