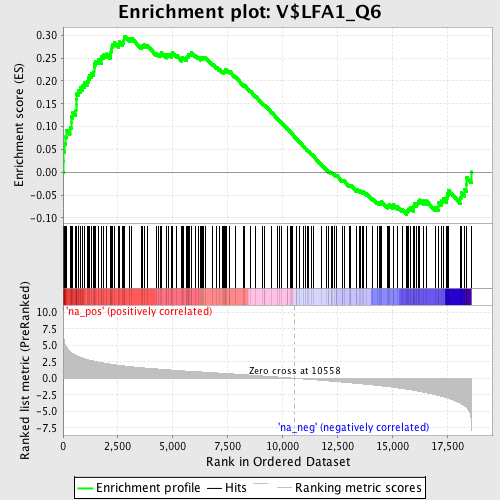
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$LFA1_Q6 |
| Enrichment Score (ES) | 0.29798457 |
| Normalized Enrichment Score (NES) | 1.4355061 |
| Nominal p-value | 0.004132231 |
| FDR q-value | 0.5009555 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MOV10 | 13 | 7.025 | 0.0240 | Yes | ||
| 2 | HES1 | 19 | 6.611 | 0.0469 | Yes | ||
| 3 | RHOG | 72 | 5.301 | 0.0627 | Yes | ||
| 4 | POLD4 | 122 | 4.910 | 0.0773 | Yes | ||
| 5 | HARS2 | 166 | 4.653 | 0.0913 | Yes | ||
| 6 | ROM1 | 314 | 4.073 | 0.0977 | Yes | ||
| 7 | TPCN1 | 369 | 3.909 | 0.1085 | Yes | ||
| 8 | PALM | 374 | 3.899 | 0.1220 | Yes | ||
| 9 | GSS | 439 | 3.726 | 0.1316 | Yes | ||
| 10 | RASGRP2 | 582 | 3.467 | 0.1361 | Yes | ||
| 11 | PDLIM7 | 588 | 3.461 | 0.1480 | Yes | ||
| 12 | SESN3 | 591 | 3.456 | 0.1600 | Yes | ||
| 13 | NCDN | 603 | 3.437 | 0.1715 | Yes | ||
| 14 | VASP | 682 | 3.311 | 0.1789 | Yes | ||
| 15 | RAB4B | 775 | 3.173 | 0.1850 | Yes | ||
| 16 | COX7A2L | 885 | 3.053 | 0.1899 | Yes | ||
| 17 | HIRA | 973 | 2.964 | 0.1956 | Yes | ||
| 18 | TITF1 | 1092 | 2.839 | 0.1991 | Yes | ||
| 19 | NEUROG1 | 1138 | 2.802 | 0.2065 | Yes | ||
| 20 | DCTN1 | 1214 | 2.738 | 0.2121 | Yes | ||
| 21 | EFNA3 | 1297 | 2.680 | 0.2171 | Yes | ||
| 22 | SEMA6A | 1403 | 2.598 | 0.2205 | Yes | ||
| 23 | CNN2 | 1408 | 2.594 | 0.2294 | Yes | ||
| 24 | PLEC1 | 1430 | 2.570 | 0.2373 | Yes | ||
| 25 | MAZ | 1484 | 2.534 | 0.2433 | Yes | ||
| 26 | UQCRC1 | 1592 | 2.462 | 0.2462 | Yes | ||
| 27 | GABRG2 | 1739 | 2.367 | 0.2466 | Yes | ||
| 28 | TRIP10 | 1752 | 2.359 | 0.2542 | Yes | ||
| 29 | MNT | 1854 | 2.299 | 0.2568 | Yes | ||
| 30 | MCRS1 | 1960 | 2.238 | 0.2590 | Yes | ||
| 31 | EPHB2 | 2147 | 2.145 | 0.2564 | Yes | ||
| 32 | SH3BP1 | 2168 | 2.133 | 0.2628 | Yes | ||
| 33 | BACE1 | 2185 | 2.127 | 0.2694 | Yes | ||
| 34 | CLIC5 | 2227 | 2.113 | 0.2746 | Yes | ||
| 35 | PDYN | 2255 | 2.096 | 0.2805 | Yes | ||
| 36 | OBSCN | 2334 | 2.057 | 0.2835 | Yes | ||
| 37 | CACNA1G | 2541 | 1.972 | 0.2793 | Yes | ||
| 38 | RABGGTA | 2558 | 1.968 | 0.2853 | Yes | ||
| 39 | DLGAP4 | 2724 | 1.892 | 0.2830 | Yes | ||
| 40 | SALL1 | 2767 | 1.878 | 0.2874 | Yes | ||
| 41 | CIC | 2775 | 1.875 | 0.2936 | Yes | ||
| 42 | HOXC6 | 2815 | 1.856 | 0.2980 | Yes | ||
| 43 | BAX | 3033 | 1.770 | 0.2924 | No | ||
| 44 | ELF4 | 3133 | 1.739 | 0.2932 | No | ||
| 45 | SLC25A29 | 3563 | 1.608 | 0.2756 | No | ||
| 46 | FKBP2 | 3629 | 1.588 | 0.2776 | No | ||
| 47 | SOX5 | 3699 | 1.569 | 0.2794 | No | ||
| 48 | SLC13A5 | 3826 | 1.530 | 0.2779 | No | ||
| 49 | KIF1C | 4257 | 1.421 | 0.2596 | No | ||
| 50 | SLC26A9 | 4366 | 1.390 | 0.2586 | No | ||
| 51 | SMAD3 | 4451 | 1.369 | 0.2589 | No | ||
| 52 | RHO | 4481 | 1.360 | 0.2621 | No | ||
| 53 | NRGN | 4704 | 1.308 | 0.2547 | No | ||
| 54 | EPO | 4723 | 1.304 | 0.2583 | No | ||
| 55 | STOML2 | 4807 | 1.284 | 0.2583 | No | ||
| 56 | TNRC4 | 4943 | 1.250 | 0.2554 | No | ||
| 57 | ADAMTS4 | 4959 | 1.245 | 0.2589 | No | ||
| 58 | USH1G | 4978 | 1.240 | 0.2623 | No | ||
| 59 | LMOD3 | 5168 | 1.195 | 0.2562 | No | ||
| 60 | ARID4A | 5408 | 1.137 | 0.2473 | No | ||
| 61 | MSX1 | 5418 | 1.135 | 0.2508 | No | ||
| 62 | SLC7A10 | 5505 | 1.114 | 0.2500 | No | ||
| 63 | LAG3 | 5613 | 1.093 | 0.2481 | No | ||
| 64 | POU4F1 | 5636 | 1.089 | 0.2507 | No | ||
| 65 | KCNAB1 | 5648 | 1.086 | 0.2539 | No | ||
| 66 | PPP1R16A | 5701 | 1.074 | 0.2549 | No | ||
| 67 | NR5A2 | 5712 | 1.072 | 0.2581 | No | ||
| 68 | NR4A1 | 5772 | 1.060 | 0.2586 | No | ||
| 69 | SIX4 | 5830 | 1.048 | 0.2592 | No | ||
| 70 | FLT1 | 5852 | 1.044 | 0.2617 | No | ||
| 71 | MRPL40 | 6030 | 1.010 | 0.2557 | No | ||
| 72 | BOC | 6171 | 0.978 | 0.2515 | No | ||
| 73 | RFX4 | 6282 | 0.956 | 0.2489 | No | ||
| 74 | IFRD2 | 6300 | 0.951 | 0.2514 | No | ||
| 75 | OTOP2 | 6370 | 0.935 | 0.2509 | No | ||
| 76 | ZFP36 | 6416 | 0.925 | 0.2517 | No | ||
| 77 | HOXC10 | 6478 | 0.914 | 0.2516 | No | ||
| 78 | ZNF385 | 6815 | 0.847 | 0.2364 | No | ||
| 79 | HNRPR | 7002 | 0.811 | 0.2291 | No | ||
| 80 | ZFPM1 | 7117 | 0.786 | 0.2257 | No | ||
| 81 | CEBPB | 7271 | 0.756 | 0.2201 | No | ||
| 82 | GBA2 | 7330 | 0.742 | 0.2195 | No | ||
| 83 | ACCN1 | 7361 | 0.737 | 0.2205 | No | ||
| 84 | GRIN2A | 7385 | 0.732 | 0.2218 | No | ||
| 85 | PTRF | 7392 | 0.730 | 0.2241 | No | ||
| 86 | DPF2 | 7442 | 0.719 | 0.2239 | No | ||
| 87 | LCAT | 7567 | 0.692 | 0.2196 | No | ||
| 88 | CREB3L2 | 7585 | 0.687 | 0.2211 | No | ||
| 89 | CNKSR2 | 7838 | 0.636 | 0.2097 | No | ||
| 90 | NDUFS2 | 8236 | 0.549 | 0.1901 | No | ||
| 91 | PNOC | 8255 | 0.544 | 0.1910 | No | ||
| 92 | TBR1 | 8530 | 0.483 | 0.1779 | No | ||
| 93 | GPR3 | 8790 | 0.422 | 0.1653 | No | ||
| 94 | GPR4 | 9102 | 0.345 | 0.1497 | No | ||
| 95 | GABARAP | 9176 | 0.328 | 0.1469 | No | ||
| 96 | S100A5 | 9197 | 0.322 | 0.1469 | No | ||
| 97 | MYBPH | 9519 | 0.258 | 0.1304 | No | ||
| 98 | KCNQ4 | 9779 | 0.196 | 0.1171 | No | ||
| 99 | PAK1IP1 | 9843 | 0.180 | 0.1143 | No | ||
| 100 | XYLT1 | 9933 | 0.156 | 0.1100 | No | ||
| 101 | CAV1 | 10226 | 0.085 | 0.0945 | No | ||
| 102 | OLFML2A | 10231 | 0.083 | 0.0945 | No | ||
| 103 | SP7 | 10376 | 0.044 | 0.0869 | No | ||
| 104 | HEBP1 | 10418 | 0.033 | 0.0848 | No | ||
| 105 | CTGF | 10454 | 0.024 | 0.0830 | No | ||
| 106 | TAL1 | 10618 | -0.014 | 0.0742 | No | ||
| 107 | KLK9 | 10650 | -0.022 | 0.0726 | No | ||
| 108 | WNT4 | 10751 | -0.050 | 0.0673 | No | ||
| 109 | JPH4 | 10969 | -0.104 | 0.0559 | No | ||
| 110 | FXYD3 | 11032 | -0.119 | 0.0530 | No | ||
| 111 | DUOX2 | 11151 | -0.149 | 0.0471 | No | ||
| 112 | PLA2G10 | 11203 | -0.163 | 0.0449 | No | ||
| 113 | NEURL | 11325 | -0.194 | 0.0390 | No | ||
| 114 | CASQ2 | 11393 | -0.209 | 0.0361 | No | ||
| 115 | PPY | 11783 | -0.311 | 0.0161 | No | ||
| 116 | NGFB | 12010 | -0.373 | 0.0052 | No | ||
| 117 | ACVR1C | 12108 | -0.398 | 0.0013 | No | ||
| 118 | CHX10 | 12114 | -0.400 | 0.0025 | No | ||
| 119 | LIN28 | 12211 | -0.424 | -0.0013 | No | ||
| 120 | NPTXR | 12243 | -0.431 | -0.0014 | No | ||
| 121 | ADM | 12284 | -0.444 | -0.0020 | No | ||
| 122 | LZTS2 | 12362 | -0.466 | -0.0046 | No | ||
| 123 | PHF12 | 12465 | -0.493 | -0.0084 | No | ||
| 124 | SMARCD2 | 12467 | -0.494 | -0.0067 | No | ||
| 125 | BNC2 | 12716 | -0.569 | -0.0181 | No | ||
| 126 | MYOG | 12754 | -0.578 | -0.0181 | No | ||
| 127 | DLX1 | 12805 | -0.592 | -0.0187 | No | ||
| 128 | CACNB1 | 13049 | -0.657 | -0.0296 | No | ||
| 129 | MGEA5 | 13080 | -0.669 | -0.0289 | No | ||
| 130 | NKX2-3 | 13112 | -0.679 | -0.0282 | No | ||
| 131 | NFKBIA | 13366 | -0.761 | -0.0392 | No | ||
| 132 | SLC2A4 | 13368 | -0.761 | -0.0366 | No | ||
| 133 | IL1F10 | 13518 | -0.803 | -0.0419 | No | ||
| 134 | NPAS3 | 13553 | -0.815 | -0.0408 | No | ||
| 135 | TSP50 | 13666 | -0.852 | -0.0439 | No | ||
| 136 | PAX7 | 13696 | -0.860 | -0.0425 | No | ||
| 137 | NFAT5 | 13837 | -0.904 | -0.0469 | No | ||
| 138 | PRRX1 | 14115 | -0.999 | -0.0584 | No | ||
| 139 | NPC2 | 14318 | -1.070 | -0.0656 | No | ||
| 140 | PYGO2 | 14419 | -1.107 | -0.0671 | No | ||
| 141 | CRMP1 | 14482 | -1.128 | -0.0665 | No | ||
| 142 | CPNE1 | 14488 | -1.129 | -0.0628 | No | ||
| 143 | AKT3 | 14774 | -1.238 | -0.0739 | No | ||
| 144 | EDG1 | 14819 | -1.256 | -0.0719 | No | ||
| 145 | THRAP1 | 14875 | -1.280 | -0.0704 | No | ||
| 146 | DHH | 15036 | -1.355 | -0.0743 | No | ||
| 147 | PCBP2 | 15050 | -1.361 | -0.0702 | No | ||
| 148 | ETV1 | 15245 | -1.448 | -0.0757 | No | ||
| 149 | GAD1 | 15447 | -1.537 | -0.0812 | No | ||
| 150 | FES | 15654 | -1.634 | -0.0866 | No | ||
| 151 | UPF2 | 15687 | -1.648 | -0.0825 | No | ||
| 152 | LRP8 | 15756 | -1.683 | -0.0803 | No | ||
| 153 | CLTC | 15811 | -1.715 | -0.0772 | No | ||
| 154 | ANXA4 | 15967 | -1.807 | -0.0793 | No | ||
| 155 | FLI1 | 15984 | -1.817 | -0.0738 | No | ||
| 156 | RAB5C | 16013 | -1.843 | -0.0688 | No | ||
| 157 | NACA | 16111 | -1.889 | -0.0674 | No | ||
| 158 | NYX | 16175 | -1.928 | -0.0641 | No | ||
| 159 | BRPF1 | 16238 | -1.962 | -0.0605 | No | ||
| 160 | RBBP6 | 16408 | -2.077 | -0.0624 | No | ||
| 161 | CKB | 16555 | -2.181 | -0.0627 | No | ||
| 162 | EMILIN1 | 16962 | -2.468 | -0.0760 | No | ||
| 163 | GNAO1 | 17109 | -2.590 | -0.0748 | No | ||
| 164 | NXF1 | 17111 | -2.591 | -0.0658 | No | ||
| 165 | CCNL1 | 17232 | -2.713 | -0.0627 | No | ||
| 166 | TCF7 | 17315 | -2.788 | -0.0574 | No | ||
| 167 | IMMT | 17483 | -2.934 | -0.0561 | No | ||
| 168 | ARID1A | 17500 | -2.949 | -0.0466 | No | ||
| 169 | GSK3B | 17578 | -3.046 | -0.0401 | No | ||
| 170 | UBL3 | 18117 | -3.780 | -0.0560 | No | ||
| 171 | CXCR4 | 18137 | -3.818 | -0.0436 | No | ||
| 172 | BCL11B | 18292 | -4.156 | -0.0374 | No | ||
| 173 | BCL6 | 18391 | -4.442 | -0.0271 | No | ||
| 174 | RBM12 | 18397 | -4.466 | -0.0117 | No | ||
| 175 | M6PR | 18604 | -6.683 | 0.0007 | No |
