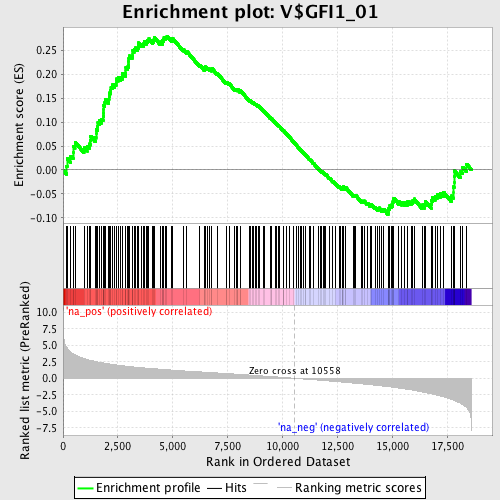
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$GFI1_01 |
| Enrichment Score (ES) | 0.2793271 |
| Normalized Enrichment Score (NES) | 1.3352517 |
| Nominal p-value | 0.033898305 |
| FDR q-value | 0.53045136 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AGPAT4 | 164 | 4.657 | 0.0080 | Yes | ||
| 2 | HMGA1 | 184 | 4.595 | 0.0236 | Yes | ||
| 3 | MAP2K5 | 350 | 3.966 | 0.0290 | Yes | ||
| 4 | BRUNOL4 | 456 | 3.697 | 0.0367 | Yes | ||
| 5 | USP5 | 463 | 3.687 | 0.0498 | Yes | ||
| 6 | BCL9L | 542 | 3.539 | 0.0584 | Yes | ||
| 7 | TCTA | 954 | 2.985 | 0.0469 | Yes | ||
| 8 | PDLIM1 | 1093 | 2.838 | 0.0497 | Yes | ||
| 9 | PRKAG1 | 1192 | 2.756 | 0.0543 | Yes | ||
| 10 | PTHLH | 1236 | 2.723 | 0.0619 | Yes | ||
| 11 | NRXN3 | 1252 | 2.712 | 0.0709 | Yes | ||
| 12 | CAMK2D | 1481 | 2.535 | 0.0677 | Yes | ||
| 13 | CACNB2 | 1500 | 2.522 | 0.0759 | Yes | ||
| 14 | GPR17 | 1505 | 2.517 | 0.0848 | Yes | ||
| 15 | RPS6KB1 | 1566 | 2.477 | 0.0905 | Yes | ||
| 16 | CLMN | 1587 | 2.467 | 0.0983 | Yes | ||
| 17 | SP8 | 1664 | 2.417 | 0.1030 | Yes | ||
| 18 | HOXA10 | 1762 | 2.351 | 0.1062 | Yes | ||
| 19 | MRGPRF | 1824 | 2.321 | 0.1113 | Yes | ||
| 20 | HOXB3 | 1837 | 2.314 | 0.1191 | Yes | ||
| 21 | HIVEP3 | 1846 | 2.305 | 0.1270 | Yes | ||
| 22 | CREB5 | 1852 | 2.299 | 0.1351 | Yes | ||
| 23 | ANGPT1 | 1903 | 2.268 | 0.1406 | Yes | ||
| 24 | HOXB8 | 1922 | 2.261 | 0.1478 | Yes | ||
| 25 | ADD3 | 2084 | 2.177 | 0.1469 | Yes | ||
| 26 | TSGA10 | 2093 | 2.169 | 0.1544 | Yes | ||
| 27 | ELAVL4 | 2120 | 2.156 | 0.1608 | Yes | ||
| 28 | BTG1 | 2162 | 2.135 | 0.1663 | Yes | ||
| 29 | ATP5G2 | 2181 | 2.128 | 0.1730 | Yes | ||
| 30 | WDR23 | 2238 | 2.106 | 0.1776 | Yes | ||
| 31 | MAP4K2 | 2357 | 2.046 | 0.1786 | Yes | ||
| 32 | CALN1 | 2414 | 2.027 | 0.1829 | Yes | ||
| 33 | HOXC8 | 2420 | 2.025 | 0.1900 | Yes | ||
| 34 | PPP2R2A | 2503 | 1.987 | 0.1927 | Yes | ||
| 35 | HIST1H1E | 2630 | 1.932 | 0.1929 | Yes | ||
| 36 | ICAM4 | 2693 | 1.906 | 0.1964 | Yes | ||
| 37 | ASB4 | 2712 | 1.899 | 0.2023 | Yes | ||
| 38 | HIST1H1T | 2839 | 1.845 | 0.2022 | Yes | ||
| 39 | CDX2 | 2860 | 1.837 | 0.2078 | Yes | ||
| 40 | DPF3 | 2862 | 1.836 | 0.2144 | Yes | ||
| 41 | GRK6 | 2954 | 1.797 | 0.2159 | Yes | ||
| 42 | KLF12 | 2985 | 1.785 | 0.2208 | Yes | ||
| 43 | RORA | 2990 | 1.783 | 0.2270 | Yes | ||
| 44 | RAB6B | 2998 | 1.780 | 0.2331 | Yes | ||
| 45 | EBF2 | 3025 | 1.772 | 0.2381 | Yes | ||
| 46 | NEXN | 3144 | 1.736 | 0.2380 | Yes | ||
| 47 | NR2F2 | 3177 | 1.727 | 0.2425 | Yes | ||
| 48 | PHACTR3 | 3179 | 1.727 | 0.2487 | Yes | ||
| 49 | ARHGEF2 | 3233 | 1.710 | 0.2520 | Yes | ||
| 50 | GPRC5D | 3297 | 1.692 | 0.2548 | Yes | ||
| 51 | TRAF4 | 3412 | 1.659 | 0.2546 | Yes | ||
| 52 | HOXB6 | 3424 | 1.651 | 0.2600 | Yes | ||
| 53 | RKHD3 | 3438 | 1.647 | 0.2652 | Yes | ||
| 54 | CPEB3 | 3594 | 1.598 | 0.2626 | Yes | ||
| 55 | SOSTDC1 | 3680 | 1.575 | 0.2637 | Yes | ||
| 56 | SOX5 | 3699 | 1.569 | 0.2684 | Yes | ||
| 57 | GFRA3 | 3809 | 1.534 | 0.2681 | Yes | ||
| 58 | PLCD4 | 3856 | 1.523 | 0.2711 | Yes | ||
| 59 | FOXP2 | 3912 | 1.508 | 0.2736 | Yes | ||
| 60 | MAP3K3 | 4089 | 1.464 | 0.2693 | Yes | ||
| 61 | ETS1 | 4141 | 1.449 | 0.2718 | Yes | ||
| 62 | DCX | 4151 | 1.447 | 0.2766 | Yes | ||
| 63 | EXOC7 | 4417 | 1.377 | 0.2672 | Yes | ||
| 64 | IL16 | 4522 | 1.352 | 0.2664 | Yes | ||
| 65 | PHYHIP | 4535 | 1.349 | 0.2707 | Yes | ||
| 66 | NFIA | 4585 | 1.334 | 0.2728 | Yes | ||
| 67 | HOXA9 | 4597 | 1.331 | 0.2771 | Yes | ||
| 68 | BDNF | 4682 | 1.313 | 0.2773 | Yes | ||
| 69 | PCDH8 | 4732 | 1.302 | 0.2793 | Yes | ||
| 70 | TCF1 | 4945 | 1.250 | 0.2724 | No | ||
| 71 | CCR7 | 4990 | 1.238 | 0.2744 | No | ||
| 72 | JMJD1C | 5477 | 1.120 | 0.2521 | No | ||
| 73 | BCL9 | 5642 | 1.087 | 0.2472 | No | ||
| 74 | MTMR4 | 6236 | 0.967 | 0.2185 | No | ||
| 75 | CHRM1 | 6442 | 0.921 | 0.2107 | No | ||
| 76 | DSG3 | 6448 | 0.920 | 0.2138 | No | ||
| 77 | RHOA | 6482 | 0.912 | 0.2153 | No | ||
| 78 | SOX9 | 6596 | 0.892 | 0.2124 | No | ||
| 79 | ZIC1 | 6689 | 0.874 | 0.2106 | No | ||
| 80 | SOX15 | 6767 | 0.857 | 0.2095 | No | ||
| 81 | IRF2BP1 | 6779 | 0.855 | 0.2120 | No | ||
| 82 | CREM | 7022 | 0.807 | 0.2018 | No | ||
| 83 | DDR2 | 7432 | 0.722 | 0.1822 | No | ||
| 84 | HOXC4 | 7463 | 0.714 | 0.1832 | No | ||
| 85 | CDON | 7566 | 0.692 | 0.1801 | No | ||
| 86 | HOXA3 | 7829 | 0.638 | 0.1682 | No | ||
| 87 | BMP4 | 7888 | 0.625 | 0.1673 | No | ||
| 88 | SLC26A3 | 7936 | 0.615 | 0.1670 | No | ||
| 89 | GREM1 | 7954 | 0.612 | 0.1683 | No | ||
| 90 | C1QTNF7 | 8072 | 0.585 | 0.1641 | No | ||
| 91 | GADD45A | 8078 | 0.584 | 0.1659 | No | ||
| 92 | HNRPA0 | 8493 | 0.494 | 0.1452 | No | ||
| 93 | TBR1 | 8530 | 0.483 | 0.1450 | No | ||
| 94 | HOXA6 | 8632 | 0.460 | 0.1412 | No | ||
| 95 | PURA | 8668 | 0.451 | 0.1410 | No | ||
| 96 | SREBF2 | 8762 | 0.427 | 0.1375 | No | ||
| 97 | PI15 | 8794 | 0.421 | 0.1373 | No | ||
| 98 | DSPP | 8884 | 0.398 | 0.1339 | No | ||
| 99 | CTLA4 | 8947 | 0.384 | 0.1319 | No | ||
| 100 | MAP2K7 | 9128 | 0.337 | 0.1234 | No | ||
| 101 | GABARAP | 9176 | 0.328 | 0.1220 | No | ||
| 102 | ITSN1 | 9430 | 0.277 | 0.1093 | No | ||
| 103 | HOXD10 | 9500 | 0.262 | 0.1065 | No | ||
| 104 | EDN1 | 9510 | 0.260 | 0.1070 | No | ||
| 105 | NKX2-8 | 9671 | 0.219 | 0.0991 | No | ||
| 106 | RKHD1 | 9747 | 0.203 | 0.0957 | No | ||
| 107 | CDH6 | 9810 | 0.188 | 0.0931 | No | ||
| 108 | BPGM | 9868 | 0.173 | 0.0906 | No | ||
| 109 | POU3F1 | 10029 | 0.135 | 0.0824 | No | ||
| 110 | MSR1 | 10044 | 0.132 | 0.0821 | No | ||
| 111 | CACNG3 | 10061 | 0.128 | 0.0817 | No | ||
| 112 | GRPR | 10178 | 0.097 | 0.0758 | No | ||
| 113 | ZFPM2 | 10318 | 0.059 | 0.0684 | No | ||
| 114 | PHF8 | 10497 | 0.012 | 0.0588 | No | ||
| 115 | PROCR | 10654 | -0.023 | 0.0504 | No | ||
| 116 | EVA1 | 10734 | -0.046 | 0.0463 | No | ||
| 117 | SERTAD4 | 10832 | -0.073 | 0.0413 | No | ||
| 118 | ZIC4 | 10877 | -0.081 | 0.0392 | No | ||
| 119 | JPH4 | 10969 | -0.104 | 0.0347 | No | ||
| 120 | CNTN6 | 11040 | -0.123 | 0.0313 | No | ||
| 121 | TLR1 | 11069 | -0.129 | 0.0303 | No | ||
| 122 | HOXD11 | 11237 | -0.172 | 0.0218 | No | ||
| 123 | SGK2 | 11282 | -0.184 | 0.0201 | No | ||
| 124 | MEIS1 | 11402 | -0.211 | 0.0144 | No | ||
| 125 | GAS2 | 11647 | -0.273 | 0.0021 | No | ||
| 126 | TRDN | 11738 | -0.296 | -0.0017 | No | ||
| 127 | RARB | 11797 | -0.316 | -0.0037 | No | ||
| 128 | PBX1 | 11859 | -0.333 | -0.0058 | No | ||
| 129 | OTP | 11913 | -0.345 | -0.0074 | No | ||
| 130 | PEA15 | 11975 | -0.360 | -0.0094 | No | ||
| 131 | MEF2C | 12142 | -0.406 | -0.0169 | No | ||
| 132 | IRX3 | 12295 | -0.445 | -0.0236 | No | ||
| 133 | KLF15 | 12415 | -0.480 | -0.0283 | No | ||
| 134 | AVP | 12574 | -0.525 | -0.0350 | No | ||
| 135 | HOXA7 | 12623 | -0.539 | -0.0356 | No | ||
| 136 | BNC2 | 12716 | -0.569 | -0.0385 | No | ||
| 137 | SH3BGRL2 | 12753 | -0.577 | -0.0384 | No | ||
| 138 | CRYZL1 | 12755 | -0.578 | -0.0364 | No | ||
| 139 | PREP | 12761 | -0.581 | -0.0345 | No | ||
| 140 | CGN | 12883 | -0.611 | -0.0389 | No | ||
| 141 | CSAD | 12890 | -0.614 | -0.0370 | No | ||
| 142 | SPAG7 | 13232 | -0.716 | -0.0529 | No | ||
| 143 | XPO5 | 13293 | -0.737 | -0.0535 | No | ||
| 144 | ABCF2 | 13333 | -0.749 | -0.0529 | No | ||
| 145 | PTEN | 13608 | -0.829 | -0.0648 | No | ||
| 146 | TFEC | 13665 | -0.852 | -0.0647 | No | ||
| 147 | NGEF | 13723 | -0.869 | -0.0647 | No | ||
| 148 | LAMA3 | 13875 | -0.917 | -0.0695 | No | ||
| 149 | PANK1 | 14014 | -0.962 | -0.0735 | No | ||
| 150 | DEPDC2 | 14039 | -0.971 | -0.0713 | No | ||
| 151 | BAI3 | 14220 | -1.034 | -0.0773 | No | ||
| 152 | PPARGC1B | 14350 | -1.079 | -0.0804 | No | ||
| 153 | TFB1M | 14401 | -1.102 | -0.0791 | No | ||
| 154 | MYO1C | 14532 | -1.147 | -0.0820 | No | ||
| 155 | PPARGC1A | 14606 | -1.174 | -0.0817 | No | ||
| 156 | TPBG | 14812 | -1.254 | -0.0883 | No | ||
| 157 | LUZP1 | 14840 | -1.268 | -0.0852 | No | ||
| 158 | SLC25A18 | 14846 | -1.269 | -0.0809 | No | ||
| 159 | ICAM5 | 14892 | -1.286 | -0.0786 | No | ||
| 160 | DSCR1L1 | 14894 | -1.287 | -0.0740 | No | ||
| 161 | TCF4 | 14964 | -1.316 | -0.0730 | No | ||
| 162 | MAP4K3 | 15024 | -1.348 | -0.0713 | No | ||
| 163 | ELMO3 | 15031 | -1.352 | -0.0668 | No | ||
| 164 | PDE4D | 15041 | -1.357 | -0.0623 | No | ||
| 165 | TRIM23 | 15074 | -1.369 | -0.0591 | No | ||
| 166 | PRIMA1 | 15306 | -1.475 | -0.0663 | No | ||
| 167 | CDH13 | 15436 | -1.530 | -0.0677 | No | ||
| 168 | ATF2 | 15554 | -1.588 | -0.0683 | No | ||
| 169 | TMPRSS2 | 15680 | -1.646 | -0.0692 | No | ||
| 170 | CNKSR1 | 15712 | -1.664 | -0.0648 | No | ||
| 171 | GPR20 | 15862 | -1.740 | -0.0666 | No | ||
| 172 | MCTS1 | 15943 | -1.792 | -0.0644 | No | ||
| 173 | MYH10 | 15992 | -1.822 | -0.0604 | No | ||
| 174 | DNAJB4 | 16359 | -2.049 | -0.0729 | No | ||
| 175 | SOX4 | 16485 | -2.128 | -0.0719 | No | ||
| 176 | ZIC3 | 16516 | -2.160 | -0.0657 | No | ||
| 177 | PDC | 16777 | -2.328 | -0.0714 | No | ||
| 178 | STMN1 | 16796 | -2.349 | -0.0639 | No | ||
| 179 | TCF12 | 16830 | -2.371 | -0.0571 | No | ||
| 180 | TOP1 | 16952 | -2.462 | -0.0547 | No | ||
| 181 | YWHAE | 17045 | -2.534 | -0.0505 | No | ||
| 182 | HSD11B1 | 17185 | -2.668 | -0.0484 | No | ||
| 183 | NCAM1 | 17330 | -2.800 | -0.0461 | No | ||
| 184 | HBP1 | 17690 | -3.155 | -0.0541 | No | ||
| 185 | POLH | 17777 | -3.265 | -0.0470 | No | ||
| 186 | SFRS2 | 17780 | -3.269 | -0.0352 | No | ||
| 187 | MBNL1 | 17821 | -3.320 | -0.0254 | No | ||
| 188 | ITGA10 | 17827 | -3.326 | -0.0136 | No | ||
| 189 | BRD2 | 17834 | -3.336 | -0.0018 | No | ||
| 190 | TLK1 | 18106 | -3.763 | -0.0029 | No | ||
| 191 | SLC4A7 | 18208 | -3.959 | 0.0060 | No | ||
| 192 | BCL6 | 18391 | -4.442 | 0.0122 | No |
