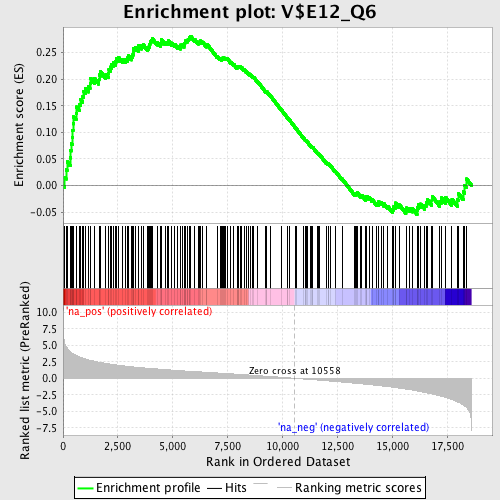
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$E12_Q6 |
| Enrichment Score (ES) | 0.28124547 |
| Normalized Enrichment Score (NES) | 1.3314025 |
| Nominal p-value | 0.03816794 |
| FDR q-value | 0.52021307 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HEYL | 82 | 5.205 | 0.0150 | Yes | ||
| 2 | AIG1 | 136 | 4.821 | 0.0302 | Yes | ||
| 3 | HMGA1 | 184 | 4.595 | 0.0449 | Yes | ||
| 4 | PTK2B | 324 | 4.044 | 0.0525 | Yes | ||
| 5 | MFNG | 340 | 3.997 | 0.0666 | Yes | ||
| 6 | ELMO1 | 371 | 3.902 | 0.0796 | Yes | ||
| 7 | ARID5A | 421 | 3.760 | 0.0910 | Yes | ||
| 8 | ENO3 | 433 | 3.738 | 0.1044 | Yes | ||
| 9 | BRUNOL4 | 456 | 3.697 | 0.1170 | Yes | ||
| 10 | ELOVL1 | 480 | 3.660 | 0.1295 | Yes | ||
| 11 | GNAS | 594 | 3.452 | 0.1363 | Yes | ||
| 12 | SASH1 | 614 | 3.408 | 0.1480 | Yes | ||
| 13 | GFRA1 | 741 | 3.221 | 0.1532 | Yes | ||
| 14 | CORO2B | 790 | 3.154 | 0.1624 | Yes | ||
| 15 | TPM3 | 898 | 3.040 | 0.1680 | Yes | ||
| 16 | PPOX | 941 | 2.998 | 0.1770 | Yes | ||
| 17 | DEF6 | 1027 | 2.895 | 0.1832 | Yes | ||
| 18 | KCNN3 | 1161 | 2.778 | 0.1864 | Yes | ||
| 19 | SLC37A4 | 1229 | 2.727 | 0.1929 | Yes | ||
| 20 | HES6 | 1259 | 2.707 | 0.2015 | Yes | ||
| 21 | PLEC1 | 1430 | 2.570 | 0.2019 | Yes | ||
| 22 | HOXB7 | 1635 | 2.438 | 0.2000 | Yes | ||
| 23 | POFUT1 | 1647 | 2.428 | 0.2085 | Yes | ||
| 24 | USP2 | 1695 | 2.392 | 0.2149 | Yes | ||
| 25 | PPP1R9B | 1951 | 2.244 | 0.2094 | Yes | ||
| 26 | SELL | 2071 | 2.185 | 0.2112 | Yes | ||
| 27 | CENTG3 | 2086 | 2.176 | 0.2185 | Yes | ||
| 28 | MTUS1 | 2149 | 2.144 | 0.2232 | Yes | ||
| 29 | CDH2 | 2199 | 2.124 | 0.2285 | Yes | ||
| 30 | IRX6 | 2280 | 2.083 | 0.2319 | Yes | ||
| 31 | PSTPIP1 | 2396 | 2.032 | 0.2333 | Yes | ||
| 32 | CALN1 | 2414 | 2.027 | 0.2400 | Yes | ||
| 33 | SIPA1 | 2536 | 1.973 | 0.2408 | Yes | ||
| 34 | GJB2 | 2721 | 1.894 | 0.2379 | Yes | ||
| 35 | DNMT3A | 2847 | 1.842 | 0.2380 | Yes | ||
| 36 | SYT4 | 2929 | 1.810 | 0.2404 | Yes | ||
| 37 | AP1S2 | 2981 | 1.787 | 0.2443 | Yes | ||
| 38 | ITGB4 | 3131 | 1.741 | 0.2428 | Yes | ||
| 39 | PHACTR3 | 3179 | 1.727 | 0.2467 | Yes | ||
| 40 | ACTN3 | 3201 | 1.721 | 0.2520 | Yes | ||
| 41 | CALB1 | 3212 | 1.716 | 0.2579 | Yes | ||
| 42 | PIM2 | 3289 | 1.695 | 0.2601 | Yes | ||
| 43 | CENTB1 | 3431 | 1.649 | 0.2586 | Yes | ||
| 44 | NTRK3 | 3447 | 1.644 | 0.2640 | Yes | ||
| 45 | CRELD1 | 3579 | 1.603 | 0.2629 | Yes | ||
| 46 | TNNC2 | 3648 | 1.583 | 0.2651 | Yes | ||
| 47 | RANGAP1 | 3852 | 1.524 | 0.2598 | Yes | ||
| 48 | GGN | 3913 | 1.508 | 0.2622 | Yes | ||
| 49 | CKM | 3949 | 1.499 | 0.2659 | Yes | ||
| 50 | COL4A3 | 3963 | 1.495 | 0.2708 | Yes | ||
| 51 | FGF8 | 4009 | 1.484 | 0.2739 | Yes | ||
| 52 | PACSIN3 | 4052 | 1.473 | 0.2771 | Yes | ||
| 53 | CYP26A1 | 4290 | 1.411 | 0.2696 | Yes | ||
| 54 | SMAD3 | 4451 | 1.369 | 0.2660 | Yes | ||
| 55 | PRKAG2 | 4467 | 1.364 | 0.2703 | Yes | ||
| 56 | CORO2A | 4482 | 1.360 | 0.2746 | Yes | ||
| 57 | GDPD2 | 4657 | 1.318 | 0.2701 | Yes | ||
| 58 | DOC2A | 4754 | 1.296 | 0.2698 | Yes | ||
| 59 | TTC17 | 4793 | 1.289 | 0.2726 | Yes | ||
| 60 | LRP5 | 4956 | 1.246 | 0.2684 | Yes | ||
| 61 | CCNL2 | 5080 | 1.216 | 0.2663 | Yes | ||
| 62 | ROBO3 | 5235 | 1.175 | 0.2624 | Yes | ||
| 63 | CNNM2 | 5338 | 1.154 | 0.2612 | Yes | ||
| 64 | IGFBP5 | 5355 | 1.150 | 0.2646 | Yes | ||
| 65 | ALDH1A2 | 5426 | 1.132 | 0.2650 | Yes | ||
| 66 | FGF11 | 5511 | 1.112 | 0.2646 | Yes | ||
| 67 | FSCN2 | 5522 | 1.110 | 0.2683 | Yes | ||
| 68 | NPEPPS | 5556 | 1.104 | 0.2706 | Yes | ||
| 69 | NOL4 | 5593 | 1.096 | 0.2727 | Yes | ||
| 70 | ARHGEF12 | 5650 | 1.086 | 0.2738 | Yes | ||
| 71 | MDGA1 | 5691 | 1.079 | 0.2756 | Yes | ||
| 72 | ARHGAP24 | 5737 | 1.067 | 0.2772 | Yes | ||
| 73 | NR4A1 | 5772 | 1.060 | 0.2793 | Yes | ||
| 74 | TNFAIP1 | 5810 | 1.051 | 0.2812 | Yes | ||
| 75 | CXCL14 | 6008 | 1.014 | 0.2744 | No | ||
| 76 | STC2 | 6160 | 0.980 | 0.2698 | No | ||
| 77 | SOST | 6217 | 0.969 | 0.2704 | No | ||
| 78 | ODC1 | 6248 | 0.964 | 0.2724 | No | ||
| 79 | CHST9 | 6354 | 0.940 | 0.2702 | No | ||
| 80 | LRP10 | 6533 | 0.903 | 0.2640 | No | ||
| 81 | CPZ | 6551 | 0.899 | 0.2664 | No | ||
| 82 | SCG2 | 7028 | 0.806 | 0.2436 | No | ||
| 83 | EYA3 | 7155 | 0.781 | 0.2397 | No | ||
| 84 | RTN4RL2 | 7239 | 0.764 | 0.2381 | No | ||
| 85 | POU2F3 | 7255 | 0.760 | 0.2401 | No | ||
| 86 | FOXI1 | 7303 | 0.747 | 0.2403 | No | ||
| 87 | GFAP | 7359 | 0.737 | 0.2401 | No | ||
| 88 | TEAD4 | 7422 | 0.724 | 0.2395 | No | ||
| 89 | ARVCF | 7481 | 0.711 | 0.2390 | No | ||
| 90 | PLCB3 | 7646 | 0.676 | 0.2326 | No | ||
| 91 | TREX2 | 7777 | 0.650 | 0.2280 | No | ||
| 92 | TCF2 | 7929 | 0.616 | 0.2221 | No | ||
| 93 | CDH5 | 7945 | 0.613 | 0.2236 | No | ||
| 94 | IGSF8 | 7989 | 0.602 | 0.2235 | No | ||
| 95 | NEGR1 | 8013 | 0.598 | 0.2245 | No | ||
| 96 | MYH14 | 8064 | 0.586 | 0.2240 | No | ||
| 97 | MYOZ3 | 8137 | 0.571 | 0.2222 | No | ||
| 98 | IGF2 | 8245 | 0.547 | 0.2185 | No | ||
| 99 | GRIA1 | 8349 | 0.523 | 0.2148 | No | ||
| 100 | MT3 | 8469 | 0.498 | 0.2103 | No | ||
| 101 | SFTPC | 8527 | 0.485 | 0.2090 | No | ||
| 102 | KLHL13 | 8636 | 0.459 | 0.2048 | No | ||
| 103 | EIF2C1 | 8680 | 0.448 | 0.2042 | No | ||
| 104 | ANK2 | 8865 | 0.402 | 0.1957 | No | ||
| 105 | DLL3 | 9240 | 0.314 | 0.1766 | No | ||
| 106 | GPM6B | 9258 | 0.312 | 0.1768 | No | ||
| 107 | ZNF205 | 9278 | 0.307 | 0.1770 | No | ||
| 108 | ANKRD2 | 9446 | 0.273 | 0.1689 | No | ||
| 109 | PTPRJ | 9931 | 0.157 | 0.1433 | No | ||
| 110 | COL4A4 | 10242 | 0.081 | 0.1268 | No | ||
| 111 | GPD1 | 10243 | 0.080 | 0.1271 | No | ||
| 112 | HMGN2 | 10306 | 0.061 | 0.1239 | No | ||
| 113 | LAMA5 | 10603 | -0.011 | 0.1079 | No | ||
| 114 | PPARA | 10656 | -0.023 | 0.1052 | No | ||
| 115 | DNAJB5 | 10940 | -0.096 | 0.0902 | No | ||
| 116 | SOX17 | 11046 | -0.124 | 0.0850 | No | ||
| 117 | BMP6 | 11052 | -0.126 | 0.0852 | No | ||
| 118 | CHRND | 11088 | -0.132 | 0.0838 | No | ||
| 119 | NBL1 | 11132 | -0.145 | 0.0820 | No | ||
| 120 | ASB14 | 11291 | -0.187 | 0.0741 | No | ||
| 121 | NEURL | 11325 | -0.194 | 0.0730 | No | ||
| 122 | TGFB3 | 11359 | -0.201 | 0.0720 | No | ||
| 123 | NHLH1 | 11374 | -0.205 | 0.0720 | No | ||
| 124 | CPA4 | 11604 | -0.261 | 0.0606 | No | ||
| 125 | DLL1 | 11645 | -0.272 | 0.0594 | No | ||
| 126 | CFL1 | 11683 | -0.281 | 0.0585 | No | ||
| 127 | NTRK2 | 11990 | -0.366 | 0.0432 | No | ||
| 128 | GATA2 | 12074 | -0.390 | 0.0402 | No | ||
| 129 | CHRDL1 | 12082 | -0.391 | 0.0413 | No | ||
| 130 | RAB26 | 12176 | -0.415 | 0.0378 | No | ||
| 131 | WTAP | 12405 | -0.476 | 0.0272 | No | ||
| 132 | SSH2 | 12718 | -0.569 | 0.0124 | No | ||
| 133 | TNFRSF21 | 13300 | -0.741 | -0.0163 | No | ||
| 134 | GPR37 | 13316 | -0.745 | -0.0143 | No | ||
| 135 | PCDH15 | 13386 | -0.766 | -0.0152 | No | ||
| 136 | RASSF2 | 13406 | -0.772 | -0.0133 | No | ||
| 137 | SLC24A3 | 13575 | -0.822 | -0.0194 | No | ||
| 138 | TNFSF13 | 13610 | -0.830 | -0.0181 | No | ||
| 139 | HSD11B2 | 13802 | -0.891 | -0.0251 | No | ||
| 140 | CANX | 13806 | -0.893 | -0.0220 | No | ||
| 141 | SORCS2 | 13830 | -0.901 | -0.0198 | No | ||
| 142 | PLAGL2 | 13943 | -0.940 | -0.0224 | No | ||
| 143 | SLC12A5 | 14092 | -0.990 | -0.0267 | No | ||
| 144 | WDR20 | 14299 | -1.064 | -0.0339 | No | ||
| 145 | GRB7 | 14378 | -1.093 | -0.0340 | No | ||
| 146 | KCNK12 | 14389 | -1.098 | -0.0305 | No | ||
| 147 | MTSS1 | 14497 | -1.132 | -0.0320 | No | ||
| 148 | FUT8 | 14622 | -1.181 | -0.0343 | No | ||
| 149 | KCNB1 | 14791 | -1.244 | -0.0388 | No | ||
| 150 | INVS | 15005 | -1.336 | -0.0453 | No | ||
| 151 | CAMK2A | 15042 | -1.358 | -0.0422 | No | ||
| 152 | TRIM23 | 15074 | -1.369 | -0.0388 | No | ||
| 153 | HR | 15148 | -1.401 | -0.0375 | No | ||
| 154 | EGLN1 | 15158 | -1.405 | -0.0327 | No | ||
| 155 | ITGA6 | 15328 | -1.484 | -0.0363 | No | ||
| 156 | WNT6 | 15629 | -1.620 | -0.0465 | No | ||
| 157 | UNC13B | 15660 | -1.636 | -0.0420 | No | ||
| 158 | SIM1 | 15786 | -1.702 | -0.0424 | No | ||
| 159 | NFATC4 | 15924 | -1.780 | -0.0432 | No | ||
| 160 | RIN1 | 16133 | -1.905 | -0.0474 | No | ||
| 161 | RAPGEF4 | 16142 | -1.911 | -0.0406 | No | ||
| 162 | FYN | 16193 | -1.940 | -0.0361 | No | ||
| 163 | POLR1D | 16306 | -2.008 | -0.0346 | No | ||
| 164 | CAMK1D | 16492 | -2.132 | -0.0367 | No | ||
| 165 | RUNX1 | 16545 | -2.175 | -0.0314 | No | ||
| 166 | GRB2 | 16609 | -2.219 | -0.0265 | No | ||
| 167 | LNX2 | 16810 | -2.359 | -0.0285 | No | ||
| 168 | DSCAM | 16814 | -2.361 | -0.0198 | No | ||
| 169 | HIF3A | 17171 | -2.652 | -0.0292 | No | ||
| 170 | SEMA3F | 17245 | -2.723 | -0.0230 | No | ||
| 171 | ADAM12 | 17441 | -2.898 | -0.0227 | No | ||
| 172 | SLC23A2 | 17721 | -3.193 | -0.0259 | No | ||
| 173 | NUP88 | 17976 | -3.558 | -0.0263 | No | ||
| 174 | RRAGC | 18027 | -3.639 | -0.0154 | No | ||
| 175 | SORT1 | 18236 | -4.021 | -0.0117 | No | ||
| 176 | OSR1 | 18294 | -4.163 | 0.0008 | No | ||
| 177 | NEDD4L | 18386 | -4.428 | 0.0125 | No |
