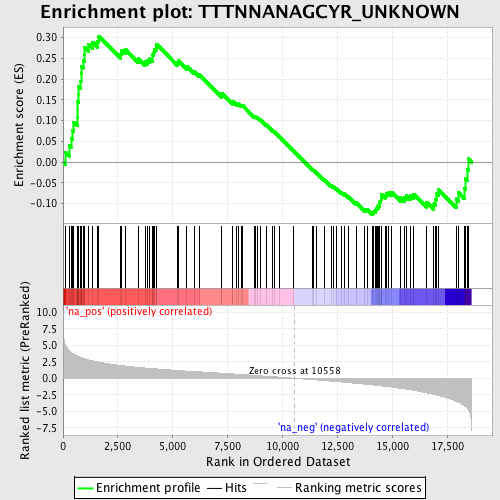
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TTTNNANAGCYR_UNKNOWN |
| Enrichment Score (ES) | 0.30375478 |
| Normalized Enrichment Score (NES) | 1.3279756 |
| Nominal p-value | 0.05323194 |
| FDR q-value | 0.5085512 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | HIST1H3C | 115 | 4.945 | 0.0232 | Yes | ||
| 2 | HIST1H2AB | 277 | 4.205 | 0.0394 | Yes | ||
| 3 | HIST1H3D | 362 | 3.933 | 0.0583 | Yes | ||
| 4 | HIST2H2BE | 441 | 3.724 | 0.0762 | Yes | ||
| 5 | HIST1H3B | 475 | 3.676 | 0.0962 | Yes | ||
| 6 | NFATC1 | 639 | 3.371 | 0.1074 | Yes | ||
| 7 | HIST1H1B | 657 | 3.342 | 0.1264 | Yes | ||
| 8 | HIST1H2BH | 658 | 3.341 | 0.1462 | Yes | ||
| 9 | HIST1H1A | 717 | 3.253 | 0.1624 | Yes | ||
| 10 | HIST1H2BN | 720 | 3.248 | 0.1816 | Yes | ||
| 11 | HIST1H2BL | 811 | 3.136 | 0.1953 | Yes | ||
| 12 | HIST1H2BJ | 817 | 3.130 | 0.2136 | Yes | ||
| 13 | HIST1H2BK | 836 | 3.102 | 0.2311 | Yes | ||
| 14 | HIST1H2BF | 915 | 3.022 | 0.2448 | Yes | ||
| 15 | INPPL1 | 967 | 2.969 | 0.2597 | Yes | ||
| 16 | HIST1H3H | 985 | 2.956 | 0.2763 | Yes | ||
| 17 | HIST1H4A | 1147 | 2.794 | 0.2842 | Yes | ||
| 18 | HIST1H2BB | 1352 | 2.629 | 0.2888 | Yes | ||
| 19 | CLDN10 | 1569 | 2.475 | 0.2919 | Yes | ||
| 20 | DPT | 1619 | 2.446 | 0.3038 | Yes | ||
| 21 | HIST1H1E | 2630 | 1.932 | 0.2607 | No | ||
| 22 | HIST1H2BA | 2675 | 1.914 | 0.2697 | No | ||
| 23 | HIST1H1T | 2839 | 1.845 | 0.2718 | No | ||
| 24 | HOXB6 | 3424 | 1.651 | 0.2501 | No | ||
| 25 | BMPR2 | 3735 | 1.557 | 0.2426 | No | ||
| 26 | RANGAP1 | 3852 | 1.524 | 0.2454 | No | ||
| 27 | HIST1H2BG | 3955 | 1.497 | 0.2488 | No | ||
| 28 | HIST1H2BC | 4071 | 1.470 | 0.2513 | No | ||
| 29 | GRK5 | 4072 | 1.469 | 0.2600 | No | ||
| 30 | ETS1 | 4141 | 1.449 | 0.2650 | No | ||
| 31 | AGTR2 | 4160 | 1.444 | 0.2726 | No | ||
| 32 | HIST1H3F | 4236 | 1.426 | 0.2770 | No | ||
| 33 | SERPINA7 | 4252 | 1.422 | 0.2846 | No | ||
| 34 | AMOT | 5198 | 1.187 | 0.2406 | No | ||
| 35 | SLC35A2 | 5240 | 1.175 | 0.2454 | No | ||
| 36 | HIST3H2BB | 5645 | 1.087 | 0.2300 | No | ||
| 37 | MYH2 | 5965 | 1.023 | 0.2189 | No | ||
| 38 | CNN1 | 6203 | 0.970 | 0.2118 | No | ||
| 39 | DHRS3 | 7222 | 0.767 | 0.1614 | No | ||
| 40 | NAP1L5 | 7230 | 0.766 | 0.1656 | No | ||
| 41 | OLR1 | 7703 | 0.666 | 0.1441 | No | ||
| 42 | DNAJB8 | 7717 | 0.664 | 0.1473 | No | ||
| 43 | DSC1 | 7903 | 0.621 | 0.1410 | No | ||
| 44 | KCNIP4 | 7980 | 0.604 | 0.1405 | No | ||
| 45 | DGKG | 8108 | 0.577 | 0.1370 | No | ||
| 46 | HIST1H2AI | 8176 | 0.562 | 0.1368 | No | ||
| 47 | SMO | 8727 | 0.436 | 0.1097 | No | ||
| 48 | CDH16 | 8753 | 0.431 | 0.1109 | No | ||
| 49 | HRASLS | 8846 | 0.408 | 0.1083 | No | ||
| 50 | NRAP | 9001 | 0.370 | 0.1022 | No | ||
| 51 | GPM6B | 9258 | 0.312 | 0.0902 | No | ||
| 52 | TCERG1L | 9552 | 0.249 | 0.0759 | No | ||
| 53 | PRKCQ | 9621 | 0.231 | 0.0736 | No | ||
| 54 | CENTD1 | 9860 | 0.176 | 0.0618 | No | ||
| 55 | FGF17 | 10516 | 0.009 | 0.0264 | No | ||
| 56 | HIST1H2AE | 11375 | -0.205 | -0.0187 | No | ||
| 57 | MEIS1 | 11402 | -0.211 | -0.0188 | No | ||
| 58 | RGN | 11548 | -0.246 | -0.0252 | No | ||
| 59 | OTP | 11913 | -0.345 | -0.0428 | No | ||
| 60 | LIN28 | 12211 | -0.424 | -0.0563 | No | ||
| 61 | FBXO21 | 12317 | -0.450 | -0.0593 | No | ||
| 62 | HIST1H2AA | 12453 | -0.489 | -0.0637 | No | ||
| 63 | HIST2H2AC | 12685 | -0.559 | -0.0728 | No | ||
| 64 | LHX2 | 12819 | -0.596 | -0.0765 | No | ||
| 65 | BLNK | 13006 | -0.643 | -0.0827 | No | ||
| 66 | SLC10A2 | 13354 | -0.758 | -0.0970 | No | ||
| 67 | ONECUT2 | 13748 | -0.878 | -0.1130 | No | ||
| 68 | HIST1H1C | 13865 | -0.913 | -0.1138 | No | ||
| 69 | NDRG3 | 14091 | -0.989 | -0.1201 | No | ||
| 70 | TBXAS1 | 14158 | -1.015 | -0.1176 | No | ||
| 71 | HIST1H1D | 14227 | -1.035 | -0.1151 | No | ||
| 72 | HIST1H2AH | 14261 | -1.048 | -0.1107 | No | ||
| 73 | NDNL2 | 14312 | -1.069 | -0.1070 | No | ||
| 74 | KCNK12 | 14389 | -1.098 | -0.1046 | No | ||
| 75 | HIST1H2BE | 14438 | -1.116 | -0.1006 | No | ||
| 76 | PPP3CA | 14441 | -1.117 | -0.0941 | No | ||
| 77 | CPNE1 | 14488 | -1.129 | -0.0898 | No | ||
| 78 | HIST3H2A | 14494 | -1.132 | -0.0834 | No | ||
| 79 | MTSS1 | 14497 | -1.132 | -0.0768 | No | ||
| 80 | HOXB4 | 14676 | -1.204 | -0.0792 | No | ||
| 81 | SPRED1 | 14736 | -1.223 | -0.0752 | No | ||
| 82 | FGD4 | 14848 | -1.270 | -0.0736 | No | ||
| 83 | TCF4 | 14964 | -1.316 | -0.0720 | No | ||
| 84 | CLDN2 | 15376 | -1.509 | -0.0852 | No | ||
| 85 | HIST1H2AC | 15546 | -1.586 | -0.0849 | No | ||
| 86 | USP47 | 15653 | -1.633 | -0.0810 | No | ||
| 87 | CAST | 15816 | -1.718 | -0.0795 | No | ||
| 88 | SLC6A1 | 15962 | -1.805 | -0.0766 | No | ||
| 89 | E2F3 | 16574 | -2.193 | -0.0966 | No | ||
| 90 | STARD13 | 16903 | -2.426 | -0.0999 | No | ||
| 91 | CYP51A1 | 16981 | -2.483 | -0.0893 | No | ||
| 92 | SLC6A13 | 17015 | -2.509 | -0.0762 | No | ||
| 93 | AOC2 | 17110 | -2.590 | -0.0659 | No | ||
| 94 | FOXP1 | 17911 | -3.457 | -0.0886 | No | ||
| 95 | ACN9 | 18019 | -3.623 | -0.0728 | No | ||
| 96 | CHD4 | 18302 | -4.175 | -0.0632 | No | ||
| 97 | MXI1 | 18336 | -4.258 | -0.0397 | No | ||
| 98 | SLC12A2 | 18413 | -4.524 | -0.0170 | No | ||
| 99 | RNF44 | 18469 | -4.696 | 0.0079 | No |
