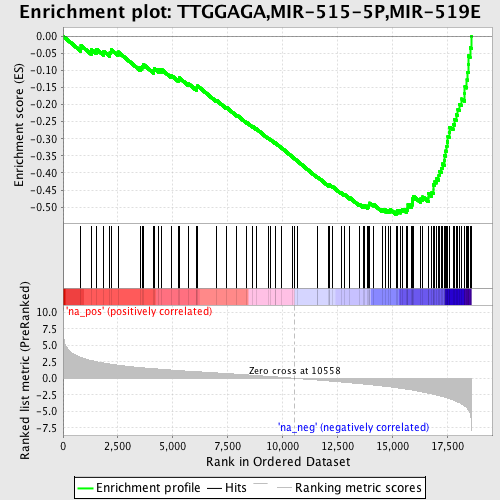
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TTGGAGA,MIR-515-5P,MIR-519E |
| Enrichment Score (ES) | -0.5215828 |
| Normalized Enrichment Score (NES) | -2.3093863 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DGCR2 | 813 | 3.133 | -0.0274 | No | ||
| 2 | EFNA3 | 1297 | 2.680 | -0.0393 | No | ||
| 3 | TGFBR2 | 1508 | 2.516 | -0.0374 | No | ||
| 4 | GABBR1 | 1853 | 2.299 | -0.0438 | No | ||
| 5 | PSMD11 | 2136 | 2.148 | -0.0477 | No | ||
| 6 | CLSTN2 | 2182 | 2.128 | -0.0389 | No | ||
| 7 | CSNK1G1 | 2505 | 1.987 | -0.0458 | No | ||
| 8 | SCN8A | 3521 | 1.622 | -0.0921 | No | ||
| 9 | NUP35 | 3596 | 1.597 | -0.0876 | No | ||
| 10 | SERTAD1 | 3655 | 1.580 | -0.0824 | No | ||
| 11 | ETS1 | 4141 | 1.449 | -0.1010 | No | ||
| 12 | DCX | 4151 | 1.447 | -0.0938 | No | ||
| 13 | FGFR2 | 4367 | 1.390 | -0.0981 | No | ||
| 14 | HOXA1 | 4500 | 1.357 | -0.0980 | No | ||
| 15 | KCND2 | 4938 | 1.252 | -0.1150 | No | ||
| 16 | POM121 | 5257 | 1.171 | -0.1260 | No | ||
| 17 | MLLT6 | 5283 | 1.164 | -0.1212 | No | ||
| 18 | GDI1 | 5694 | 1.077 | -0.1377 | No | ||
| 19 | SLC4A4 | 6084 | 0.996 | -0.1534 | No | ||
| 20 | SLC9A2 | 6097 | 0.993 | -0.1489 | No | ||
| 21 | APLN | 6127 | 0.987 | -0.1452 | No | ||
| 22 | SOX30 | 6998 | 0.811 | -0.1879 | No | ||
| 23 | FSTL1 | 7439 | 0.721 | -0.2079 | No | ||
| 24 | CPEB2 | 7919 | 0.619 | -0.2305 | No | ||
| 25 | CLCF1 | 8363 | 0.519 | -0.2517 | No | ||
| 26 | NFIB | 8635 | 0.460 | -0.2639 | No | ||
| 27 | MEF2D | 8804 | 0.420 | -0.2707 | No | ||
| 28 | KCTD7 | 9358 | 0.290 | -0.2991 | No | ||
| 29 | ITSN1 | 9430 | 0.277 | -0.3014 | No | ||
| 30 | ACY1L2 | 9664 | 0.219 | -0.3129 | No | ||
| 31 | ABCC3 | 9670 | 0.219 | -0.3120 | No | ||
| 32 | CAB39 | 9966 | 0.150 | -0.3271 | No | ||
| 33 | MTF2 | 10461 | 0.021 | -0.3537 | No | ||
| 34 | SP4 | 10550 | 0.002 | -0.3584 | No | ||
| 35 | ETNK2 | 10665 | -0.026 | -0.3644 | No | ||
| 36 | ANUBL1 | 11597 | -0.259 | -0.4134 | No | ||
| 37 | UBE2B | 11616 | -0.263 | -0.4129 | No | ||
| 38 | SNAP25 | 12104 | -0.397 | -0.4372 | No | ||
| 39 | BSN | 12128 | -0.403 | -0.4363 | No | ||
| 40 | EMX2 | 12129 | -0.403 | -0.4341 | No | ||
| 41 | ISL1 | 12288 | -0.444 | -0.4403 | No | ||
| 42 | ARRDC3 | 12676 | -0.555 | -0.4583 | No | ||
| 43 | MOSPD2 | 12830 | -0.599 | -0.4634 | No | ||
| 44 | SYNGR1 | 13073 | -0.666 | -0.4730 | No | ||
| 45 | TMEM47 | 13487 | -0.795 | -0.4911 | No | ||
| 46 | OSBPL3 | 13692 | -0.860 | -0.4976 | No | ||
| 47 | DDX3Y | 13721 | -0.868 | -0.4945 | No | ||
| 48 | NOTCH2 | 13889 | -0.921 | -0.4986 | No | ||
| 49 | ARHGEF3 | 13936 | -0.938 | -0.4962 | No | ||
| 50 | PLAGL2 | 13943 | -0.940 | -0.4915 | No | ||
| 51 | OPHN1 | 13970 | -0.948 | -0.4879 | No | ||
| 52 | RSBN1 | 14139 | -1.009 | -0.4917 | No | ||
| 53 | ABCG4 | 14546 | -1.156 | -0.5075 | No | ||
| 54 | DONSON | 14675 | -1.204 | -0.5081 | No | ||
| 55 | ATRN | 14830 | -1.265 | -0.5097 | No | ||
| 56 | FRAS1 | 14919 | -1.299 | -0.5076 | No | ||
| 57 | RPS6KA3 | 15179 | -1.415 | -0.5141 | Yes | ||
| 58 | ETV1 | 15245 | -1.448 | -0.5100 | Yes | ||
| 59 | SGPL1 | 15383 | -1.511 | -0.5094 | Yes | ||
| 60 | ULK2 | 15470 | -1.548 | -0.5059 | Yes | ||
| 61 | BTG2 | 15657 | -1.634 | -0.5073 | Yes | ||
| 62 | ATXN3 | 15715 | -1.664 | -0.5016 | Yes | ||
| 63 | PDGFRA | 15717 | -1.665 | -0.4929 | Yes | ||
| 64 | IQGAP1 | 15888 | -1.760 | -0.4927 | Yes | ||
| 65 | SOX12 | 15912 | -1.770 | -0.4846 | Yes | ||
| 66 | NFATC4 | 15924 | -1.780 | -0.4758 | Yes | ||
| 67 | EPN2 | 15959 | -1.803 | -0.4682 | Yes | ||
| 68 | CYR61 | 16302 | -2.003 | -0.4761 | Yes | ||
| 69 | MTHFD1L | 16397 | -2.071 | -0.4702 | Yes | ||
| 70 | POU2F1 | 16649 | -2.246 | -0.4719 | Yes | ||
| 71 | KPNA1 | 16651 | -2.248 | -0.4601 | Yes | ||
| 72 | HIP2 | 16808 | -2.357 | -0.4561 | Yes | ||
| 73 | HNRPUL1 | 16880 | -2.408 | -0.4472 | Yes | ||
| 74 | NCOA1 | 16881 | -2.409 | -0.4345 | Yes | ||
| 75 | SLC7A11 | 16945 | -2.458 | -0.4249 | Yes | ||
| 76 | UBE2H | 17032 | -2.527 | -0.4162 | Yes | ||
| 77 | MARCKS | 17128 | -2.603 | -0.4076 | Yes | ||
| 78 | RDH11 | 17142 | -2.625 | -0.3944 | Yes | ||
| 79 | PUM1 | 17246 | -2.724 | -0.3856 | Yes | ||
| 80 | ZBTB1 | 17295 | -2.771 | -0.3736 | Yes | ||
| 81 | YTHDF2 | 17382 | -2.842 | -0.3632 | Yes | ||
| 82 | SUMO2 | 17392 | -2.853 | -0.3486 | Yes | ||
| 83 | TNPO1 | 17436 | -2.895 | -0.3357 | Yes | ||
| 84 | UBAP2 | 17487 | -2.938 | -0.3229 | Yes | ||
| 85 | ARID1A | 17500 | -2.949 | -0.3079 | Yes | ||
| 86 | EPC2 | 17538 | -3.007 | -0.2941 | Yes | ||
| 87 | RAP2C | 17595 | -3.061 | -0.2809 | Yes | ||
| 88 | CMTM6 | 17622 | -3.085 | -0.2660 | Yes | ||
| 89 | DDX3X | 17776 | -3.265 | -0.2571 | Yes | ||
| 90 | MAPRE1 | 17831 | -3.327 | -0.2424 | Yes | ||
| 91 | YAF2 | 17919 | -3.470 | -0.2288 | Yes | ||
| 92 | RIOK3 | 17993 | -3.591 | -0.2138 | Yes | ||
| 93 | RAP1B | 18070 | -3.696 | -0.1984 | Yes | ||
| 94 | UHRF2 | 18175 | -3.890 | -0.1835 | Yes | ||
| 95 | ZFP36L1 | 18305 | -4.187 | -0.1683 | Yes | ||
| 96 | RBL2 | 18314 | -4.213 | -0.1465 | Yes | ||
| 97 | SFRS1 | 18407 | -4.486 | -0.1278 | Yes | ||
| 98 | TMEM2 | 18443 | -4.619 | -0.1053 | Yes | ||
| 99 | RPP14 | 18470 | -4.699 | -0.0819 | Yes | ||
| 100 | AP3S1 | 18479 | -4.726 | -0.0574 | Yes | ||
| 101 | ICK | 18550 | -5.291 | -0.0332 | Yes | ||
| 102 | FREQ | 18608 | -6.962 | 0.0004 | Yes |
