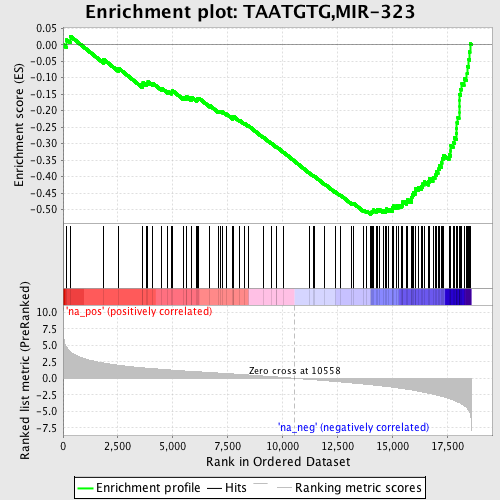
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TAATGTG,MIR-323 |
| Enrichment Score (ES) | -0.5150609 |
| Normalized Enrichment Score (NES) | -2.2701247 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SPSB4 | 159 | 4.687 | 0.0149 | No | ||
| 2 | PTK2B | 324 | 4.044 | 0.0262 | No | ||
| 3 | EGR3 | 1848 | 2.302 | -0.0445 | No | ||
| 4 | ACAT2 | 2518 | 1.983 | -0.0708 | No | ||
| 5 | ANKRD27 | 3607 | 1.594 | -0.1216 | No | ||
| 6 | TMEM16D | 3639 | 1.587 | -0.1153 | No | ||
| 7 | PAK7 | 3801 | 1.536 | -0.1163 | No | ||
| 8 | STAT5B | 3867 | 1.521 | -0.1122 | No | ||
| 9 | MAP3K3 | 4089 | 1.464 | -0.1168 | No | ||
| 10 | HOXA1 | 4500 | 1.357 | -0.1322 | No | ||
| 11 | SEPT3 | 4777 | 1.292 | -0.1406 | No | ||
| 12 | HOXB5 | 4944 | 1.250 | -0.1434 | No | ||
| 13 | OSBP | 4962 | 1.244 | -0.1380 | No | ||
| 14 | STK35 | 5473 | 1.121 | -0.1600 | No | ||
| 15 | RAB11FIP2 | 5608 | 1.094 | -0.1618 | No | ||
| 16 | CDKN1B | 5634 | 1.090 | -0.1577 | No | ||
| 17 | FBN1 | 5838 | 1.047 | -0.1634 | No | ||
| 18 | KCNJ3 | 5863 | 1.042 | -0.1595 | No | ||
| 19 | SLC4A4 | 6084 | 0.996 | -0.1664 | No | ||
| 20 | CRK | 6133 | 0.986 | -0.1640 | No | ||
| 21 | MCFD2 | 6184 | 0.974 | -0.1619 | No | ||
| 22 | ZIC1 | 6689 | 0.874 | -0.1847 | No | ||
| 23 | CXCL12 | 7092 | 0.791 | -0.2025 | No | ||
| 24 | NFKB1 | 7163 | 0.779 | -0.2024 | No | ||
| 25 | POU2F3 | 7255 | 0.760 | -0.2035 | No | ||
| 26 | DPF2 | 7442 | 0.719 | -0.2099 | No | ||
| 27 | TMOD1 | 7742 | 0.658 | -0.2228 | No | ||
| 28 | EIF3S1 | 7768 | 0.652 | -0.2209 | No | ||
| 29 | GPM6A | 7774 | 0.651 | -0.2179 | No | ||
| 30 | STX12 | 8039 | 0.591 | -0.2292 | No | ||
| 31 | LPP | 8283 | 0.541 | -0.2396 | No | ||
| 32 | SEMA6D | 8429 | 0.506 | -0.2449 | No | ||
| 33 | RCN1 | 9129 | 0.337 | -0.2810 | No | ||
| 34 | NRK | 9479 | 0.266 | -0.2985 | No | ||
| 35 | MYLIP | 9721 | 0.208 | -0.3105 | No | ||
| 36 | FBN2 | 9733 | 0.206 | -0.3101 | No | ||
| 37 | BET1 | 10051 | 0.130 | -0.3265 | No | ||
| 38 | EBF3 | 11227 | -0.168 | -0.3892 | No | ||
| 39 | QSCN6L1 | 11394 | -0.209 | -0.3971 | No | ||
| 40 | TGFA | 11462 | -0.225 | -0.3996 | No | ||
| 41 | AUH | 11920 | -0.346 | -0.4226 | No | ||
| 42 | WTAP | 12405 | -0.476 | -0.4463 | No | ||
| 43 | TEAD1 | 12634 | -0.542 | -0.4559 | No | ||
| 44 | LRRC4 | 13154 | -0.691 | -0.4805 | No | ||
| 45 | POU4F2 | 13226 | -0.714 | -0.4808 | No | ||
| 46 | ENTPD5 | 13700 | -0.861 | -0.5020 | No | ||
| 47 | NFAT5 | 13837 | -0.904 | -0.5048 | No | ||
| 48 | ATP11B | 14027 | -0.967 | -0.5102 | Yes | ||
| 49 | CHCHD4 | 14067 | -0.981 | -0.5074 | Yes | ||
| 50 | DCUN1D1 | 14112 | -0.998 | -0.5048 | Yes | ||
| 51 | RSBN1 | 14139 | -1.009 | -0.5012 | Yes | ||
| 52 | PDCD10 | 14274 | -1.054 | -0.5031 | Yes | ||
| 53 | NR4A3 | 14316 | -1.070 | -0.5000 | Yes | ||
| 54 | CCND1 | 14405 | -1.103 | -0.4992 | Yes | ||
| 55 | PPARGC1A | 14606 | -1.174 | -0.5042 | Yes | ||
| 56 | HOOK3 | 14694 | -1.211 | -0.5028 | Yes | ||
| 57 | WBP2 | 14719 | -1.217 | -0.4980 | Yes | ||
| 58 | FGD4 | 14848 | -1.270 | -0.4986 | Yes | ||
| 59 | ASF1A | 14999 | -1.333 | -0.5000 | Yes | ||
| 60 | PCDH9 | 15000 | -1.333 | -0.4933 | Yes | ||
| 61 | PJA2 | 15046 | -1.359 | -0.4890 | Yes | ||
| 62 | RPS6KA3 | 15179 | -1.415 | -0.4890 | Yes | ||
| 63 | MAP4K4 | 15300 | -1.473 | -0.4881 | Yes | ||
| 64 | TBPL1 | 15409 | -1.519 | -0.4864 | Yes | ||
| 65 | ULK2 | 15470 | -1.548 | -0.4818 | Yes | ||
| 66 | ZDHHC21 | 15485 | -1.553 | -0.4748 | Yes | ||
| 67 | CSNK1G3 | 15661 | -1.636 | -0.4761 | Yes | ||
| 68 | TOP2B | 15703 | -1.659 | -0.4700 | Yes | ||
| 69 | RAB10 | 15871 | -1.750 | -0.4703 | Yes | ||
| 70 | CUL1 | 15887 | -1.757 | -0.4623 | Yes | ||
| 71 | PKP4 | 15908 | -1.767 | -0.4545 | Yes | ||
| 72 | SLITRK2 | 15968 | -1.808 | -0.4487 | Yes | ||
| 73 | FEM1B | 16053 | -1.857 | -0.4439 | Yes | ||
| 74 | MRC1 | 16069 | -1.866 | -0.4354 | Yes | ||
| 75 | CEPT1 | 16206 | -1.949 | -0.4330 | Yes | ||
| 76 | UTX | 16343 | -2.031 | -0.4302 | Yes | ||
| 77 | ZHX1 | 16391 | -2.067 | -0.4224 | Yes | ||
| 78 | CLCN3 | 16452 | -2.107 | -0.4151 | Yes | ||
| 79 | KPNA1 | 16651 | -2.248 | -0.4145 | Yes | ||
| 80 | KPNA3 | 16694 | -2.272 | -0.4054 | Yes | ||
| 81 | SMARCAD1 | 16860 | -2.394 | -0.4024 | Yes | ||
| 82 | TOP1 | 16952 | -2.462 | -0.3950 | Yes | ||
| 83 | USP9X | 17012 | -2.508 | -0.3856 | Yes | ||
| 84 | GJA1 | 17089 | -2.575 | -0.3768 | Yes | ||
| 85 | DHX36 | 17150 | -2.636 | -0.3669 | Yes | ||
| 86 | TNRC6A | 17242 | -2.721 | -0.3582 | Yes | ||
| 87 | CLASP1 | 17269 | -2.744 | -0.3458 | Yes | ||
| 88 | TRIM33 | 17323 | -2.795 | -0.3347 | Yes | ||
| 89 | NFIX | 17591 | -3.058 | -0.3338 | Yes | ||
| 90 | REV3L | 17668 | -3.128 | -0.3223 | Yes | ||
| 91 | CREB1 | 17673 | -3.133 | -0.3068 | Yes | ||
| 92 | DDX3X | 17776 | -3.265 | -0.2960 | Yes | ||
| 93 | MAPRE1 | 17831 | -3.327 | -0.2823 | Yes | ||
| 94 | HIPK1 | 17930 | -3.486 | -0.2701 | Yes | ||
| 95 | SLMAP | 17942 | -3.502 | -0.2532 | Yes | ||
| 96 | PPP1CB | 17950 | -3.512 | -0.2360 | Yes | ||
| 97 | ZNF318 | 17992 | -3.591 | -0.2202 | Yes | ||
| 98 | RANBP9 | 18046 | -3.662 | -0.2048 | Yes | ||
| 99 | PITPNA | 18049 | -3.673 | -0.1865 | Yes | ||
| 100 | ARFIP1 | 18052 | -3.677 | -0.1682 | Yes | ||
| 101 | RAB1A | 18056 | -3.680 | -0.1499 | Yes | ||
| 102 | UBL3 | 18117 | -3.780 | -0.1343 | Yes | ||
| 103 | UHRF2 | 18175 | -3.890 | -0.1179 | Yes | ||
| 104 | SUMO1 | 18274 | -4.114 | -0.1026 | Yes | ||
| 105 | SFRS1 | 18407 | -4.486 | -0.0873 | Yes | ||
| 106 | NDEL1 | 18422 | -4.555 | -0.0652 | Yes | ||
| 107 | TMPO | 18492 | -4.809 | -0.0449 | Yes | ||
| 108 | HS2ST1 | 18529 | -5.048 | -0.0216 | Yes | ||
| 109 | SATB1 | 18546 | -5.244 | 0.0038 | Yes |
