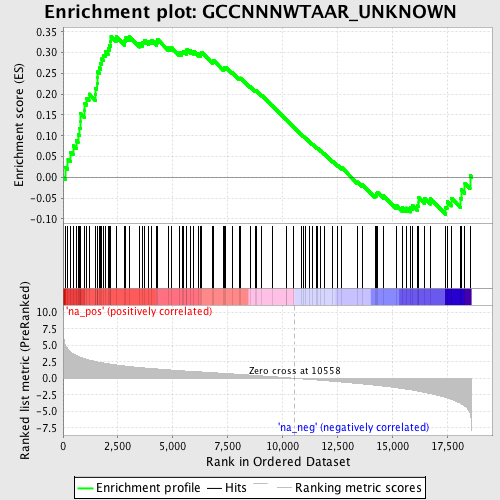
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GCCNNNWTAAR_UNKNOWN |
| Enrichment Score (ES) | 0.33995983 |
| Normalized Enrichment Score (NES) | 1.48979 |
| Nominal p-value | 0.012704174 |
| FDR q-value | 0.4065571 |
| FWER p-Value | 0.993 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RPP21 | 96 | 5.078 | 0.0246 | Yes | ||
| 2 | GRASP | 221 | 4.399 | 0.0437 | Yes | ||
| 3 | GGA1 | 342 | 3.990 | 0.0606 | Yes | ||
| 4 | BRUNOL4 | 456 | 3.697 | 0.0761 | Yes | ||
| 5 | CCBL1 | 599 | 3.444 | 0.0886 | Yes | ||
| 6 | VASP | 682 | 3.311 | 0.1036 | Yes | ||
| 7 | EGFL6 | 756 | 3.206 | 0.1185 | Yes | ||
| 8 | ETV6 | 787 | 3.156 | 0.1354 | Yes | ||
| 9 | ETV5 | 791 | 3.153 | 0.1537 | Yes | ||
| 10 | INPPL1 | 967 | 2.969 | 0.1616 | Yes | ||
| 11 | GNAQ | 974 | 2.964 | 0.1787 | Yes | ||
| 12 | CTDSP1 | 1064 | 2.863 | 0.1906 | Yes | ||
| 13 | MMP24 | 1181 | 2.762 | 0.2006 | Yes | ||
| 14 | RPL10 | 1471 | 2.541 | 0.1998 | Yes | ||
| 15 | CAMK2D | 1481 | 2.535 | 0.2142 | Yes | ||
| 16 | EGR1 | 1544 | 2.490 | 0.2255 | Yes | ||
| 17 | CDKN2C | 1556 | 2.484 | 0.2394 | Yes | ||
| 18 | SLC39A5 | 1561 | 2.481 | 0.2538 | Yes | ||
| 19 | LDHD | 1646 | 2.430 | 0.2635 | Yes | ||
| 20 | VAX1 | 1707 | 2.385 | 0.2742 | Yes | ||
| 21 | HOXA10 | 1762 | 2.351 | 0.2851 | Yes | ||
| 22 | PTPRG | 1851 | 2.301 | 0.2938 | Yes | ||
| 23 | HOXB8 | 1922 | 2.261 | 0.3033 | Yes | ||
| 24 | NKX2-5 | 2049 | 2.195 | 0.3093 | Yes | ||
| 25 | ELAVL4 | 2120 | 2.156 | 0.3182 | Yes | ||
| 26 | FHL3 | 2173 | 2.131 | 0.3279 | Yes | ||
| 27 | ATP5G2 | 2181 | 2.128 | 0.3400 | Yes | ||
| 28 | PEX5 | 2416 | 2.026 | 0.3392 | No | ||
| 29 | TBCC | 2816 | 1.856 | 0.3285 | No | ||
| 30 | DPF3 | 2862 | 1.836 | 0.3369 | No | ||
| 31 | ABCC5 | 3020 | 1.773 | 0.3388 | No | ||
| 32 | KLC2 | 3487 | 1.631 | 0.3232 | No | ||
| 33 | SLC25A10 | 3627 | 1.589 | 0.3250 | No | ||
| 34 | SOX5 | 3699 | 1.569 | 0.3303 | No | ||
| 35 | FOXP2 | 3912 | 1.508 | 0.3277 | No | ||
| 36 | H2AFX | 4030 | 1.480 | 0.3301 | No | ||
| 37 | NFIL3 | 4258 | 1.420 | 0.3261 | No | ||
| 38 | HMGA2 | 4313 | 1.407 | 0.3315 | No | ||
| 39 | RAB33A | 4818 | 1.282 | 0.3118 | No | ||
| 40 | LRP5 | 4956 | 1.246 | 0.3117 | No | ||
| 41 | NDRG2 | 5306 | 1.160 | 0.2996 | No | ||
| 42 | MSX1 | 5418 | 1.135 | 0.3003 | No | ||
| 43 | STK35 | 5473 | 1.121 | 0.3039 | No | ||
| 44 | CDKN1B | 5634 | 1.090 | 0.3017 | No | ||
| 45 | BCL9 | 5642 | 1.087 | 0.3077 | No | ||
| 46 | GDNF | 5806 | 1.052 | 0.3050 | No | ||
| 47 | PHOX2B | 5964 | 1.023 | 0.3025 | No | ||
| 48 | ASGR1 | 6166 | 0.979 | 0.2974 | No | ||
| 49 | RFX4 | 6282 | 0.956 | 0.2968 | No | ||
| 50 | COCH | 6309 | 0.949 | 0.3010 | No | ||
| 51 | HTR2C | 6797 | 0.852 | 0.2797 | No | ||
| 52 | MID1 | 6841 | 0.841 | 0.2823 | No | ||
| 53 | FGF9 | 7305 | 0.747 | 0.2616 | No | ||
| 54 | BHLHB3 | 7339 | 0.741 | 0.2642 | No | ||
| 55 | IMPDH2 | 7417 | 0.725 | 0.2643 | No | ||
| 56 | TRIB2 | 7700 | 0.667 | 0.2530 | No | ||
| 57 | PDGFB | 8041 | 0.591 | 0.2381 | No | ||
| 58 | DAB1 | 8086 | 0.582 | 0.2391 | No | ||
| 59 | SFTPC | 8527 | 0.485 | 0.2182 | No | ||
| 60 | ABCA1 | 8747 | 0.432 | 0.2089 | No | ||
| 61 | MEF2D | 8804 | 0.420 | 0.2083 | No | ||
| 62 | KCNH3 | 9043 | 0.359 | 0.1976 | No | ||
| 63 | MMP9 | 9527 | 0.256 | 0.1730 | No | ||
| 64 | ADAMTS5 | 10172 | 0.100 | 0.1388 | No | ||
| 65 | OTX1 | 10186 | 0.095 | 0.1387 | No | ||
| 66 | SFXN5 | 10483 | 0.015 | 0.1228 | No | ||
| 67 | NPR3 | 10857 | -0.077 | 0.1031 | No | ||
| 68 | JPH4 | 10969 | -0.104 | 0.0977 | No | ||
| 69 | HOXA4 | 11051 | -0.125 | 0.0940 | No | ||
| 70 | EN2 | 11218 | -0.165 | 0.0860 | No | ||
| 71 | TGFB3 | 11359 | -0.201 | 0.0797 | No | ||
| 72 | PHEX | 11550 | -0.247 | 0.0708 | No | ||
| 73 | SLC5A7 | 11588 | -0.255 | 0.0703 | No | ||
| 74 | NPTX1 | 11752 | -0.300 | 0.0633 | No | ||
| 75 | OTP | 11913 | -0.345 | 0.0567 | No | ||
| 76 | ISL1 | 12288 | -0.444 | 0.0391 | No | ||
| 77 | EML4 | 12526 | -0.510 | 0.0293 | No | ||
| 78 | KCTD15 | 12706 | -0.566 | 0.0229 | No | ||
| 79 | MITF | 13424 | -0.775 | -0.0113 | No | ||
| 80 | ATOH1 | 13639 | -0.842 | -0.0179 | No | ||
| 81 | SHH | 14216 | -1.033 | -0.0429 | No | ||
| 82 | APBA2BP | 14265 | -1.049 | -0.0394 | No | ||
| 83 | ERG | 14317 | -1.070 | -0.0359 | No | ||
| 84 | PPARGC1A | 14606 | -1.174 | -0.0445 | No | ||
| 85 | MPP2 | 15194 | -1.422 | -0.0679 | No | ||
| 86 | ITGA3 | 15457 | -1.541 | -0.0730 | No | ||
| 87 | FES | 15654 | -1.634 | -0.0740 | No | ||
| 88 | XPO7 | 15835 | -1.727 | -0.0736 | No | ||
| 89 | NFATC4 | 15924 | -1.780 | -0.0680 | No | ||
| 90 | ZADH2 | 16149 | -1.914 | -0.0688 | No | ||
| 91 | FYN | 16193 | -1.940 | -0.0598 | No | ||
| 92 | UBE2D3 | 16195 | -1.941 | -0.0485 | No | ||
| 93 | SOX4 | 16485 | -2.128 | -0.0516 | No | ||
| 94 | VCL | 16727 | -2.291 | -0.0512 | No | ||
| 95 | ADAM12 | 17441 | -2.898 | -0.0727 | No | ||
| 96 | ARID1A | 17500 | -2.949 | -0.0586 | No | ||
| 97 | SOCS2 | 17700 | -3.168 | -0.0507 | No | ||
| 98 | RBMX | 18107 | -3.763 | -0.0506 | No | ||
| 99 | PTMA | 18146 | -3.836 | -0.0302 | No | ||
| 100 | HPS3 | 18313 | -4.205 | -0.0145 | No | ||
| 101 | ITPR1 | 18547 | -5.257 | 0.0037 | No |
