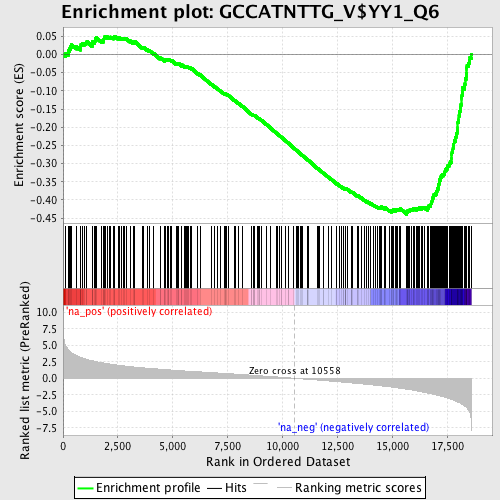
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | GCCATNTTG_V$YY1_Q6 |
| Enrichment Score (ES) | -0.4401455 |
| Normalized Enrichment Score (NES) | -2.2378879 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AIP | 123 | 4.909 | 0.0025 | No | ||
| 2 | UBE1 | 230 | 4.376 | 0.0049 | No | ||
| 3 | IDH3G | 244 | 4.335 | 0.0123 | No | ||
| 4 | AGPAT1 | 306 | 4.100 | 0.0166 | No | ||
| 5 | MAP2K5 | 350 | 3.966 | 0.0217 | No | ||
| 6 | MPDU1 | 389 | 3.858 | 0.0268 | No | ||
| 7 | NCDN | 603 | 3.437 | 0.0216 | No | ||
| 8 | RAB4B | 775 | 3.173 | 0.0182 | No | ||
| 9 | TAF6 | 808 | 3.141 | 0.0223 | No | ||
| 10 | DGCR2 | 813 | 3.133 | 0.0280 | No | ||
| 11 | RXRB | 871 | 3.073 | 0.0306 | No | ||
| 12 | USF1 | 957 | 2.983 | 0.0315 | No | ||
| 13 | NDUFS8 | 1044 | 2.883 | 0.0322 | No | ||
| 14 | WBSCR16 | 1086 | 2.842 | 0.0353 | No | ||
| 15 | SLC39A7 | 1324 | 2.656 | 0.0273 | No | ||
| 16 | PIAS3 | 1329 | 2.648 | 0.0321 | No | ||
| 17 | STRN4 | 1347 | 2.634 | 0.0361 | No | ||
| 18 | COX8A | 1449 | 2.559 | 0.0353 | No | ||
| 19 | EEF1B2 | 1459 | 2.553 | 0.0396 | No | ||
| 20 | RPL10 | 1471 | 2.541 | 0.0438 | No | ||
| 21 | CACNB2 | 1500 | 2.522 | 0.0470 | No | ||
| 22 | BTRC | 1757 | 2.357 | 0.0374 | No | ||
| 23 | LPHN1 | 1821 | 2.322 | 0.0383 | No | ||
| 24 | MNT | 1854 | 2.299 | 0.0409 | No | ||
| 25 | SMYD5 | 1863 | 2.295 | 0.0447 | No | ||
| 26 | NUDT3 | 1875 | 2.286 | 0.0484 | No | ||
| 27 | PMF1 | 1921 | 2.261 | 0.0502 | No | ||
| 28 | SRGAP2 | 2025 | 2.210 | 0.0487 | No | ||
| 29 | ELAVL4 | 2120 | 2.156 | 0.0476 | No | ||
| 30 | ATP5G2 | 2181 | 2.128 | 0.0483 | No | ||
| 31 | HIF1A | 2285 | 2.081 | 0.0465 | No | ||
| 32 | ATP6V0C | 2322 | 2.063 | 0.0484 | No | ||
| 33 | ABHD1 | 2352 | 2.050 | 0.0507 | No | ||
| 34 | PPP2R2A | 2503 | 1.987 | 0.0462 | No | ||
| 35 | CELSR3 | 2574 | 1.960 | 0.0461 | No | ||
| 36 | PPP4R2 | 2676 | 1.914 | 0.0441 | No | ||
| 37 | SNX17 | 2750 | 1.887 | 0.0437 | No | ||
| 38 | HOXC6 | 2815 | 1.856 | 0.0437 | No | ||
| 39 | RBM10 | 2877 | 1.832 | 0.0438 | No | ||
| 40 | WDR13 | 3053 | 1.763 | 0.0375 | No | ||
| 41 | UBE3A | 3207 | 1.719 | 0.0324 | No | ||
| 42 | MTMR3 | 3241 | 1.708 | 0.0338 | No | ||
| 43 | DHX35 | 3266 | 1.700 | 0.0356 | No | ||
| 44 | LYPLA2 | 3625 | 1.589 | 0.0191 | No | ||
| 45 | PITX1 | 3654 | 1.581 | 0.0205 | No | ||
| 46 | U2AF2 | 3837 | 1.528 | 0.0134 | No | ||
| 47 | RNF5 | 3930 | 1.505 | 0.0112 | No | ||
| 48 | SF3A1 | 4128 | 1.452 | 0.0032 | No | ||
| 49 | EXOC7 | 4417 | 1.377 | -0.0099 | No | ||
| 50 | APBB1 | 4452 | 1.369 | -0.0092 | No | ||
| 51 | POLR2A | 4625 | 1.325 | -0.0161 | No | ||
| 52 | EIF2B4 | 4628 | 1.324 | -0.0138 | No | ||
| 53 | FKRP | 4678 | 1.314 | -0.0140 | No | ||
| 54 | IRS4 | 4742 | 1.299 | -0.0150 | No | ||
| 55 | DYRK1B | 4783 | 1.291 | -0.0148 | No | ||
| 56 | TTC17 | 4793 | 1.289 | -0.0128 | No | ||
| 57 | ATF4 | 4881 | 1.263 | -0.0152 | No | ||
| 58 | ATRX | 4951 | 1.248 | -0.0166 | No | ||
| 59 | FZD8 | 5149 | 1.198 | -0.0252 | No | ||
| 60 | RPL18A | 5206 | 1.184 | -0.0260 | No | ||
| 61 | TAX1BP3 | 5256 | 1.171 | -0.0265 | No | ||
| 62 | SCRN1 | 5262 | 1.170 | -0.0246 | No | ||
| 63 | ARID4A | 5408 | 1.137 | -0.0303 | No | ||
| 64 | TSC1 | 5409 | 1.137 | -0.0282 | No | ||
| 65 | CHES1 | 5540 | 1.107 | -0.0332 | No | ||
| 66 | ZBTB4 | 5592 | 1.096 | -0.0340 | No | ||
| 67 | CDKN1B | 5634 | 1.090 | -0.0342 | No | ||
| 68 | MRPL11 | 5668 | 1.083 | -0.0339 | No | ||
| 69 | UQCRH | 5713 | 1.072 | -0.0343 | No | ||
| 70 | PSPC1 | 5790 | 1.055 | -0.0365 | No | ||
| 71 | AP4S1 | 5873 | 1.041 | -0.0390 | No | ||
| 72 | RAGE | 6146 | 0.983 | -0.0520 | No | ||
| 73 | L3MBTL2 | 6243 | 0.966 | -0.0555 | No | ||
| 74 | ATP5A1 | 6760 | 0.859 | -0.0820 | No | ||
| 75 | ZFP37 | 6910 | 0.827 | -0.0886 | No | ||
| 76 | APLP1 | 7046 | 0.800 | -0.0944 | No | ||
| 77 | CBX5 | 7193 | 0.773 | -0.1010 | No | ||
| 78 | A2BP1 | 7334 | 0.742 | -0.1072 | No | ||
| 79 | SHANK2 | 7380 | 0.733 | -0.1083 | No | ||
| 80 | ING4 | 7408 | 0.726 | -0.1084 | No | ||
| 81 | HOXC4 | 7463 | 0.714 | -0.1100 | No | ||
| 82 | GIT2 | 7533 | 0.699 | -0.1125 | No | ||
| 83 | DISP2 | 7809 | 0.644 | -0.1263 | No | ||
| 84 | TOP3A | 7858 | 0.631 | -0.1277 | No | ||
| 85 | SLC26A6 | 7987 | 0.603 | -0.1336 | No | ||
| 86 | CPSF2 | 8165 | 0.564 | -0.1422 | No | ||
| 87 | LMO6 | 8601 | 0.467 | -0.1650 | No | ||
| 88 | PTBP1 | 8605 | 0.467 | -0.1643 | No | ||
| 89 | RACGAP1 | 8674 | 0.449 | -0.1672 | No | ||
| 90 | UBE2J2 | 8697 | 0.445 | -0.1675 | No | ||
| 91 | ZBTB11 | 8726 | 0.437 | -0.1682 | No | ||
| 92 | ADAM11 | 8743 | 0.432 | -0.1683 | No | ||
| 93 | JARID2 | 8859 | 0.404 | -0.1738 | No | ||
| 94 | IRAK1 | 8906 | 0.393 | -0.1756 | No | ||
| 95 | ARHGEF17 | 8951 | 0.383 | -0.1773 | No | ||
| 96 | NFYC | 8966 | 0.379 | -0.1773 | No | ||
| 97 | MAEA | 9038 | 0.360 | -0.1805 | No | ||
| 98 | OTX2 | 9275 | 0.307 | -0.1928 | No | ||
| 99 | ATF1 | 9473 | 0.268 | -0.2031 | No | ||
| 100 | DUSP7 | 9743 | 0.204 | -0.2174 | No | ||
| 101 | ATP5G1 | 9791 | 0.193 | -0.2196 | No | ||
| 102 | HIC2 | 9872 | 0.172 | -0.2236 | No | ||
| 103 | RFX1 | 9947 | 0.154 | -0.2273 | No | ||
| 104 | RPS8 | 9953 | 0.153 | -0.2273 | No | ||
| 105 | BTF3 | 10116 | 0.115 | -0.2360 | No | ||
| 106 | KPNA6 | 10267 | 0.073 | -0.2440 | No | ||
| 107 | MAP3K4 | 10269 | 0.072 | -0.2439 | No | ||
| 108 | SMCR8 | 10514 | 0.009 | -0.2572 | No | ||
| 109 | PCTK2 | 10632 | -0.019 | -0.2635 | No | ||
| 110 | RPL35A | 10660 | -0.025 | -0.2650 | No | ||
| 111 | SEC23A | 10722 | -0.042 | -0.2682 | No | ||
| 112 | UBA52 | 10814 | -0.068 | -0.2731 | No | ||
| 113 | UPF3B | 10858 | -0.077 | -0.2753 | No | ||
| 114 | MYST2 | 10906 | -0.088 | -0.2777 | No | ||
| 115 | SYT11 | 11125 | -0.143 | -0.2893 | No | ||
| 116 | IRX5 | 11126 | -0.144 | -0.2890 | No | ||
| 117 | ATP5F1 | 11131 | -0.145 | -0.2890 | No | ||
| 118 | ARMET | 11178 | -0.156 | -0.2912 | No | ||
| 119 | NDUFS1 | 11605 | -0.261 | -0.3139 | No | ||
| 120 | RAB5A | 11621 | -0.265 | -0.3142 | No | ||
| 121 | WHSC2 | 11666 | -0.279 | -0.3161 | No | ||
| 122 | PAPOLB | 11877 | -0.338 | -0.3269 | No | ||
| 123 | SSR4 | 11886 | -0.339 | -0.3267 | No | ||
| 124 | UBAP1 | 12102 | -0.397 | -0.3377 | No | ||
| 125 | NR2C2 | 12229 | -0.429 | -0.3438 | No | ||
| 126 | PHF12 | 12465 | -0.493 | -0.3557 | No | ||
| 127 | TIA1 | 12471 | -0.494 | -0.3550 | No | ||
| 128 | TGM5 | 12593 | -0.530 | -0.3606 | No | ||
| 129 | UBC | 12674 | -0.553 | -0.3640 | No | ||
| 130 | PRKCSH | 12771 | -0.583 | -0.3681 | No | ||
| 131 | BOP1 | 12774 | -0.583 | -0.3671 | No | ||
| 132 | GRIN2B | 12852 | -0.602 | -0.3702 | No | ||
| 133 | RNF26 | 12866 | -0.607 | -0.3698 | No | ||
| 134 | ENSA | 12881 | -0.611 | -0.3694 | No | ||
| 135 | GFER | 12885 | -0.612 | -0.3684 | No | ||
| 136 | BAMBI | 12958 | -0.629 | -0.3712 | No | ||
| 137 | PNRC2 | 13135 | -0.684 | -0.3795 | No | ||
| 138 | LRRC4 | 13154 | -0.691 | -0.3792 | No | ||
| 139 | THAP1 | 13160 | -0.691 | -0.3782 | No | ||
| 140 | PCF11 | 13200 | -0.708 | -0.3790 | No | ||
| 141 | TARDBP | 13395 | -0.768 | -0.3881 | No | ||
| 142 | AP3D1 | 13430 | -0.777 | -0.3885 | No | ||
| 143 | PCBP4 | 13467 | -0.789 | -0.3890 | No | ||
| 144 | METTL3 | 13585 | -0.825 | -0.3938 | No | ||
| 145 | RPN1 | 13745 | -0.878 | -0.4009 | No | ||
| 146 | CHD2 | 13845 | -0.907 | -0.4046 | No | ||
| 147 | LMNB1 | 13920 | -0.934 | -0.4069 | No | ||
| 148 | CCNK | 14021 | -0.965 | -0.4105 | No | ||
| 149 | CAB39L | 14025 | -0.967 | -0.4089 | No | ||
| 150 | ACTR8 | 14165 | -1.018 | -0.4145 | No | ||
| 151 | PROSC | 14250 | -1.042 | -0.4172 | No | ||
| 152 | FUS | 14343 | -1.078 | -0.4202 | No | ||
| 153 | PPP1CC | 14433 | -1.114 | -0.4230 | No | ||
| 154 | SFRS7 | 14444 | -1.118 | -0.4214 | No | ||
| 155 | BCL2L13 | 14484 | -1.128 | -0.4214 | No | ||
| 156 | CPNE1 | 14488 | -1.129 | -0.4195 | No | ||
| 157 | KCNMA1 | 14499 | -1.134 | -0.4179 | No | ||
| 158 | PSMC5 | 14638 | -1.189 | -0.4232 | No | ||
| 159 | DOCK8 | 14666 | -1.200 | -0.4224 | No | ||
| 160 | ING3 | 14674 | -1.204 | -0.4206 | No | ||
| 161 | KNTC1 | 14880 | -1.281 | -0.4294 | No | ||
| 162 | TCF4 | 14964 | -1.316 | -0.4314 | No | ||
| 163 | KIF1B | 14985 | -1.327 | -0.4300 | No | ||
| 164 | MAP4K3 | 15024 | -1.348 | -0.4296 | No | ||
| 165 | PDE4D | 15041 | -1.357 | -0.4279 | No | ||
| 166 | PFN2 | 15068 | -1.367 | -0.4268 | No | ||
| 167 | HSF1 | 15161 | -1.405 | -0.4292 | No | ||
| 168 | RPS6KA3 | 15179 | -1.415 | -0.4275 | No | ||
| 169 | CNOT3 | 15202 | -1.430 | -0.4260 | No | ||
| 170 | ETV1 | 15245 | -1.448 | -0.4256 | No | ||
| 171 | SNX5 | 15329 | -1.484 | -0.4273 | No | ||
| 172 | AEBP2 | 15343 | -1.494 | -0.4252 | No | ||
| 173 | CASC3 | 15379 | -1.510 | -0.4243 | No | ||
| 174 | RUFY1 | 15670 | -1.641 | -0.4371 | Yes | ||
| 175 | THRAP4 | 15673 | -1.642 | -0.4341 | Yes | ||
| 176 | METTL4 | 15702 | -1.659 | -0.4325 | Yes | ||
| 177 | ELK3 | 15714 | -1.664 | -0.4300 | Yes | ||
| 178 | STRN3 | 15746 | -1.679 | -0.4286 | Yes | ||
| 179 | PAPOLG | 15798 | -1.709 | -0.4282 | Yes | ||
| 180 | RBM3 | 15809 | -1.712 | -0.4255 | Yes | ||
| 181 | RAB10 | 15871 | -1.750 | -0.4256 | Yes | ||
| 182 | COX7B | 15951 | -1.798 | -0.4265 | Yes | ||
| 183 | SLC6A1 | 15962 | -1.805 | -0.4237 | Yes | ||
| 184 | DDX6 | 16027 | -1.848 | -0.4237 | Yes | ||
| 185 | NACA | 16111 | -1.889 | -0.4247 | Yes | ||
| 186 | RAPGEF4 | 16142 | -1.911 | -0.4228 | Yes | ||
| 187 | HOXA2 | 16204 | -1.948 | -0.4225 | Yes | ||
| 188 | NSF | 16225 | -1.957 | -0.4199 | Yes | ||
| 189 | UTX | 16343 | -2.031 | -0.4225 | Yes | ||
| 190 | RAB22A | 16381 | -2.062 | -0.4207 | Yes | ||
| 191 | PRPSAP2 | 16461 | -2.113 | -0.4210 | Yes | ||
| 192 | AP1G1 | 16593 | -2.204 | -0.4241 | Yes | ||
| 193 | ZFX | 16602 | -2.215 | -0.4204 | Yes | ||
| 194 | POU2F1 | 16649 | -2.246 | -0.4187 | Yes | ||
| 195 | EIF5 | 16674 | -2.258 | -0.4158 | Yes | ||
| 196 | RBM16 | 16742 | -2.302 | -0.4151 | Yes | ||
| 197 | DAP3 | 16763 | -2.320 | -0.4119 | Yes | ||
| 198 | RNF12 | 16767 | -2.321 | -0.4077 | Yes | ||
| 199 | HIP2 | 16808 | -2.357 | -0.4055 | Yes | ||
| 200 | DSCAM | 16814 | -2.361 | -0.4013 | Yes | ||
| 201 | RRM1 | 16847 | -2.385 | -0.3986 | Yes | ||
| 202 | RAD21 | 16853 | -2.389 | -0.3944 | Yes | ||
| 203 | VIL2 | 16866 | -2.398 | -0.3906 | Yes | ||
| 204 | RALA | 16872 | -2.405 | -0.3864 | Yes | ||
| 205 | HNRPA1 | 16925 | -2.440 | -0.3847 | Yes | ||
| 206 | ARCN1 | 16978 | -2.481 | -0.3829 | Yes | ||
| 207 | SFRS3 | 17008 | -2.503 | -0.3798 | Yes | ||
| 208 | ARPC2 | 17021 | -2.516 | -0.3757 | Yes | ||
| 209 | YWHAE | 17045 | -2.534 | -0.3722 | Yes | ||
| 210 | NFE2L1 | 17066 | -2.552 | -0.3686 | Yes | ||
| 211 | MARK3 | 17098 | -2.584 | -0.3654 | Yes | ||
| 212 | GNAO1 | 17109 | -2.590 | -0.3611 | Yes | ||
| 213 | RBMS1 | 17117 | -2.594 | -0.3567 | Yes | ||
| 214 | ASXL2 | 17137 | -2.617 | -0.3528 | Yes | ||
| 215 | SFRS10 | 17151 | -2.636 | -0.3486 | Yes | ||
| 216 | CCND2 | 17169 | -2.650 | -0.3446 | Yes | ||
| 217 | AKAP8L | 17197 | -2.682 | -0.3410 | Yes | ||
| 218 | HN1 | 17215 | -2.697 | -0.3369 | Yes | ||
| 219 | PUM1 | 17246 | -2.724 | -0.3335 | Yes | ||
| 220 | USP14 | 17271 | -2.747 | -0.3296 | Yes | ||
| 221 | PPP1R15B | 17357 | -2.820 | -0.3290 | Yes | ||
| 222 | SUMO2 | 17392 | -2.853 | -0.3255 | Yes | ||
| 223 | ARF1 | 17403 | -2.867 | -0.3207 | Yes | ||
| 224 | RECK | 17413 | -2.876 | -0.3158 | Yes | ||
| 225 | NCOA6 | 17463 | -2.917 | -0.3130 | Yes | ||
| 226 | MYNN | 17504 | -2.952 | -0.3097 | Yes | ||
| 227 | EPC2 | 17538 | -3.007 | -0.3059 | Yes | ||
| 228 | APRIN | 17589 | -3.058 | -0.3029 | Yes | ||
| 229 | CSNK1A1 | 17614 | -3.081 | -0.2985 | Yes | ||
| 230 | SFXN1 | 17655 | -3.117 | -0.2948 | Yes | ||
| 231 | CD4 | 17701 | -3.168 | -0.2913 | Yes | ||
| 232 | RBM5 | 17702 | -3.169 | -0.2854 | Yes | ||
| 233 | NFYA | 17710 | -3.180 | -0.2799 | Yes | ||
| 234 | MRPL51 | 17711 | -3.181 | -0.2739 | Yes | ||
| 235 | TIAL1 | 17722 | -3.194 | -0.2685 | Yes | ||
| 236 | SFRS5 | 17737 | -3.212 | -0.2633 | Yes | ||
| 237 | TRIM8 | 17769 | -3.256 | -0.2589 | Yes | ||
| 238 | DDX3X | 17776 | -3.265 | -0.2531 | Yes | ||
| 239 | SFRS2 | 17780 | -3.269 | -0.2471 | Yes | ||
| 240 | WASL | 17815 | -3.310 | -0.2428 | Yes | ||
| 241 | PIAS1 | 17820 | -3.319 | -0.2368 | Yes | ||
| 242 | ILF3 | 17878 | -3.408 | -0.2336 | Yes | ||
| 243 | EIF4A1 | 17880 | -3.409 | -0.2273 | Yes | ||
| 244 | FOXP1 | 17911 | -3.457 | -0.2224 | Yes | ||
| 245 | CRKL | 17939 | -3.501 | -0.2174 | Yes | ||
| 246 | EIF4G2 | 17956 | -3.525 | -0.2117 | Yes | ||
| 247 | CLK4 | 17982 | -3.578 | -0.2063 | Yes | ||
| 248 | HSPA8 | 17987 | -3.581 | -0.1999 | Yes | ||
| 249 | TAF15 | 17991 | -3.590 | -0.1933 | Yes | ||
| 250 | ZNF318 | 17992 | -3.591 | -0.1866 | Yes | ||
| 251 | MRPL1 | 18016 | -3.620 | -0.1811 | Yes | ||
| 252 | SFPQ | 18040 | -3.653 | -0.1755 | Yes | ||
| 253 | ZDHHC5 | 18042 | -3.656 | -0.1687 | Yes | ||
| 254 | RAB1A | 18056 | -3.680 | -0.1626 | Yes | ||
| 255 | CD164 | 18073 | -3.702 | -0.1565 | Yes | ||
| 256 | MAT2A | 18098 | -3.752 | -0.1508 | Yes | ||
| 257 | RBMX | 18107 | -3.763 | -0.1442 | Yes | ||
| 258 | CSE1L | 18131 | -3.809 | -0.1384 | Yes | ||
| 259 | TNRC15 | 18145 | -3.835 | -0.1319 | Yes | ||
| 260 | PTMA | 18146 | -3.836 | -0.1247 | Yes | ||
| 261 | PPP2CA | 18149 | -3.839 | -0.1177 | Yes | ||
| 262 | ANAPC4 | 18177 | -3.891 | -0.1119 | Yes | ||
| 263 | HNRPC | 18191 | -3.911 | -0.1053 | Yes | ||
| 264 | AKAP8 | 18210 | -3.963 | -0.0989 | Yes | ||
| 265 | UGCGL1 | 18213 | -3.968 | -0.0916 | Yes | ||
| 266 | CUGBP1 | 18289 | -4.148 | -0.0879 | Yes | ||
| 267 | PAFAH1B1 | 18307 | -4.189 | -0.0810 | Yes | ||
| 268 | ATE1 | 18323 | -4.239 | -0.0739 | Yes | ||
| 269 | H1F0 | 18324 | -4.240 | -0.0660 | Yes | ||
| 270 | FTSJ3 | 18367 | -4.344 | -0.0602 | Yes | ||
| 271 | ADK | 18378 | -4.388 | -0.0525 | Yes | ||
| 272 | RBM12 | 18397 | -4.466 | -0.0451 | Yes | ||
| 273 | NASP | 18399 | -4.468 | -0.0368 | Yes | ||
| 274 | SFRS1 | 18407 | -4.486 | -0.0288 | Yes | ||
| 275 | CTCF | 18467 | -4.689 | -0.0233 | Yes | ||
| 276 | SP1 | 18508 | -4.895 | -0.0163 | Yes | ||
| 277 | WSB1 | 18541 | -5.191 | -0.0084 | Yes | ||
| 278 | EPC1 | 18603 | -6.642 | 0.0007 | Yes |
