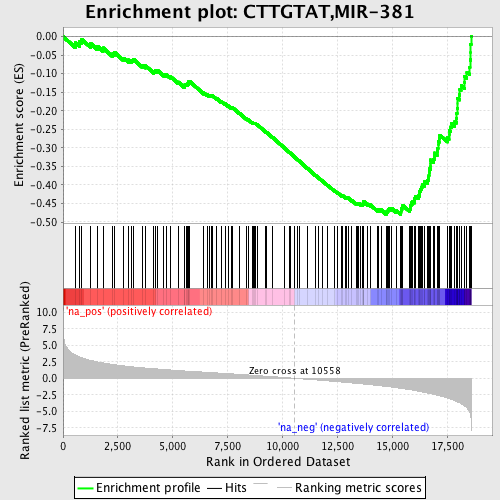
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CTTGTAT,MIR-381 |
| Enrichment Score (ES) | -0.4797397 |
| Normalized Enrichment Score (NES) | -2.25716 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DLC1 | 562 | 3.505 | -0.0167 | No | ||
| 2 | GFRA1 | 741 | 3.221 | -0.0138 | No | ||
| 3 | DHX40 | 851 | 3.092 | -0.0076 | No | ||
| 4 | AGPAT3 | 1248 | 2.716 | -0.0184 | No | ||
| 5 | RPS6KB1 | 1566 | 2.477 | -0.0259 | No | ||
| 6 | LPHN1 | 1821 | 2.322 | -0.0306 | No | ||
| 7 | ACTG1 | 2268 | 2.090 | -0.0466 | No | ||
| 8 | UBN1 | 2335 | 2.057 | -0.0421 | No | ||
| 9 | RALGPS1 | 2774 | 1.875 | -0.0585 | No | ||
| 10 | KLF12 | 2985 | 1.785 | -0.0629 | No | ||
| 11 | GRM8 | 3120 | 1.744 | -0.0634 | No | ||
| 12 | UBE3A | 3207 | 1.719 | -0.0613 | No | ||
| 13 | TMEM16D | 3639 | 1.587 | -0.0784 | No | ||
| 14 | GORASP2 | 3760 | 1.549 | -0.0789 | No | ||
| 15 | ETS1 | 4141 | 1.449 | -0.0938 | No | ||
| 16 | FLRT3 | 4209 | 1.434 | -0.0918 | No | ||
| 17 | WNT5A | 4322 | 1.403 | -0.0924 | No | ||
| 18 | NFIA | 4585 | 1.334 | -0.1014 | No | ||
| 19 | BCL11A | 4692 | 1.312 | -0.1020 | No | ||
| 20 | TRIM63 | 4879 | 1.264 | -0.1071 | No | ||
| 21 | COL3A1 | 5249 | 1.173 | -0.1225 | No | ||
| 22 | BAT2 | 5531 | 1.108 | -0.1334 | No | ||
| 23 | MDS1 | 5553 | 1.105 | -0.1302 | No | ||
| 24 | RAB11FIP2 | 5608 | 1.094 | -0.1289 | No | ||
| 25 | RNF13 | 5662 | 1.085 | -0.1275 | No | ||
| 26 | NR5A2 | 5712 | 1.072 | -0.1260 | No | ||
| 27 | VSNL1 | 5718 | 1.071 | -0.1221 | No | ||
| 28 | CACNA1C | 5771 | 1.060 | -0.1208 | No | ||
| 29 | NRXN1 | 6408 | 0.928 | -0.1516 | No | ||
| 30 | EPHA4 | 6559 | 0.898 | -0.1562 | No | ||
| 31 | ZCCHC11 | 6675 | 0.877 | -0.1590 | No | ||
| 32 | PHF15 | 6741 | 0.863 | -0.1591 | No | ||
| 33 | ACVR2A | 6808 | 0.850 | -0.1594 | No | ||
| 34 | HNRPR | 7002 | 0.811 | -0.1667 | No | ||
| 35 | NAP1L5 | 7230 | 0.766 | -0.1760 | No | ||
| 36 | SHANK2 | 7380 | 0.733 | -0.1812 | No | ||
| 37 | CRSP2 | 7537 | 0.698 | -0.1869 | No | ||
| 38 | PDGFC | 7683 | 0.669 | -0.1921 | No | ||
| 39 | SIPA1L2 | 7720 | 0.663 | -0.1915 | No | ||
| 40 | STX12 | 8039 | 0.591 | -0.2064 | No | ||
| 41 | CLCF1 | 8363 | 0.519 | -0.2219 | No | ||
| 42 | SEMA6D | 8429 | 0.506 | -0.2234 | No | ||
| 43 | PNRC1 | 8625 | 0.462 | -0.2322 | No | ||
| 44 | SRRM1 | 8679 | 0.449 | -0.2333 | No | ||
| 45 | PMP22 | 8714 | 0.442 | -0.2334 | No | ||
| 46 | ABCA1 | 8747 | 0.432 | -0.2334 | No | ||
| 47 | ST8SIA2 | 8851 | 0.406 | -0.2374 | No | ||
| 48 | TBC1D15 | 9239 | 0.314 | -0.2572 | No | ||
| 49 | EPHA3 | 9284 | 0.305 | -0.2583 | No | ||
| 50 | RNF2 | 9535 | 0.255 | -0.2709 | No | ||
| 51 | PARD6B | 10112 | 0.116 | -0.3016 | No | ||
| 52 | ZFPM2 | 10318 | 0.059 | -0.3125 | No | ||
| 53 | RHOQ | 10353 | 0.051 | -0.3141 | No | ||
| 54 | UBE2E2 | 10525 | 0.006 | -0.3234 | No | ||
| 55 | RPL35A | 10660 | -0.025 | -0.3305 | No | ||
| 56 | BIRC6 | 10752 | -0.051 | -0.3353 | No | ||
| 57 | BTBD7 | 11145 | -0.147 | -0.3559 | No | ||
| 58 | JPH1 | 11510 | -0.236 | -0.3747 | No | ||
| 59 | GAS2 | 11647 | -0.273 | -0.3810 | No | ||
| 60 | UBE4A | 11805 | -0.319 | -0.3882 | No | ||
| 61 | CCNT2 | 12065 | -0.387 | -0.4008 | No | ||
| 62 | LZTS2 | 12362 | -0.466 | -0.4150 | No | ||
| 63 | FOXF2 | 12498 | -0.503 | -0.4203 | No | ||
| 64 | KCTD15 | 12706 | -0.566 | -0.4293 | No | ||
| 65 | SSH2 | 12718 | -0.569 | -0.4277 | No | ||
| 66 | PBEF1 | 12882 | -0.611 | -0.4341 | No | ||
| 67 | PAPOLA | 12900 | -0.615 | -0.4326 | No | ||
| 68 | JAG2 | 12991 | -0.638 | -0.4350 | No | ||
| 69 | LRRC4 | 13154 | -0.691 | -0.4411 | No | ||
| 70 | NFKBIA | 13366 | -0.761 | -0.4495 | No | ||
| 71 | MITF | 13424 | -0.775 | -0.4496 | No | ||
| 72 | BHLHB5 | 13472 | -0.789 | -0.4491 | No | ||
| 73 | SLC24A3 | 13575 | -0.822 | -0.4514 | No | ||
| 74 | GRM1 | 13649 | -0.844 | -0.4520 | No | ||
| 75 | B3GNT5 | 13680 | -0.856 | -0.4503 | No | ||
| 76 | PURG | 13684 | -0.858 | -0.4471 | No | ||
| 77 | OSBPL3 | 13692 | -0.860 | -0.4441 | No | ||
| 78 | MBNL2 | 13893 | -0.923 | -0.4514 | No | ||
| 79 | ELOVL7 | 14003 | -0.959 | -0.4535 | No | ||
| 80 | QKI | 14346 | -1.078 | -0.4678 | No | ||
| 81 | ZFAND3 | 14383 | -1.094 | -0.4655 | No | ||
| 82 | MTSS1 | 14497 | -1.132 | -0.4672 | No | ||
| 83 | CGGBP1 | 14724 | -1.218 | -0.4747 | Yes | ||
| 84 | ACSL3 | 14755 | -1.230 | -0.4715 | Yes | ||
| 85 | ARID4B | 14805 | -1.250 | -0.4693 | Yes | ||
| 86 | ATRN | 14830 | -1.265 | -0.4656 | Yes | ||
| 87 | THRAP1 | 14875 | -1.280 | -0.4630 | Yes | ||
| 88 | KIF1B | 14985 | -1.327 | -0.4637 | Yes | ||
| 89 | RPS6KA3 | 15179 | -1.415 | -0.4686 | Yes | ||
| 90 | RNF144 | 15385 | -1.512 | -0.4738 | Yes | ||
| 91 | SNRK | 15401 | -1.517 | -0.4687 | Yes | ||
| 92 | R3HDM1 | 15426 | -1.527 | -0.4641 | Yes | ||
| 93 | GAD1 | 15447 | -1.537 | -0.4591 | Yes | ||
| 94 | HNRPH2 | 15482 | -1.551 | -0.4549 | Yes | ||
| 95 | PAPOLG | 15798 | -1.709 | -0.4653 | Yes | ||
| 96 | XPO7 | 15835 | -1.727 | -0.4605 | Yes | ||
| 97 | PHF2 | 15845 | -1.731 | -0.4542 | Yes | ||
| 98 | AZIN1 | 15863 | -1.740 | -0.4483 | Yes | ||
| 99 | PSIP1 | 15927 | -1.781 | -0.4448 | Yes | ||
| 100 | ITCH | 16003 | -1.837 | -0.4417 | Yes | ||
| 101 | DDX6 | 16027 | -1.848 | -0.4357 | Yes | ||
| 102 | SLC6A8 | 16058 | -1.858 | -0.4301 | Yes | ||
| 103 | KCTD3 | 16189 | -1.936 | -0.4295 | Yes | ||
| 104 | ANKRD50 | 16228 | -1.957 | -0.4240 | Yes | ||
| 105 | THRAP2 | 16247 | -1.968 | -0.4172 | Yes | ||
| 106 | YTHDF1 | 16282 | -1.989 | -0.4113 | Yes | ||
| 107 | KLF4 | 16323 | -2.019 | -0.4056 | Yes | ||
| 108 | NDUFAB1 | 16362 | -2.050 | -0.3996 | Yes | ||
| 109 | STAG1 | 16457 | -2.109 | -0.3965 | Yes | ||
| 110 | SOX4 | 16485 | -2.128 | -0.3896 | Yes | ||
| 111 | SEMA3C | 16605 | -2.215 | -0.3874 | Yes | ||
| 112 | ROCK2 | 16643 | -2.242 | -0.3807 | Yes | ||
| 113 | WEE1 | 16673 | -2.258 | -0.3734 | Yes | ||
| 114 | NLK | 16679 | -2.261 | -0.3649 | Yes | ||
| 115 | KPNA3 | 16694 | -2.272 | -0.3567 | Yes | ||
| 116 | RBM16 | 16742 | -2.302 | -0.3503 | Yes | ||
| 117 | EIF1 | 16749 | -2.308 | -0.3416 | Yes | ||
| 118 | SYNCRIP | 16752 | -2.313 | -0.3327 | Yes | ||
| 119 | NIN | 16882 | -2.410 | -0.3302 | Yes | ||
| 120 | RNF111 | 16922 | -2.437 | -0.3228 | Yes | ||
| 121 | CSTF3 | 16935 | -2.449 | -0.3139 | Yes | ||
| 122 | EIF3S10 | 17047 | -2.537 | -0.3100 | Yes | ||
| 123 | PDAP1 | 17075 | -2.562 | -0.3015 | Yes | ||
| 124 | GJA1 | 17089 | -2.575 | -0.2921 | Yes | ||
| 125 | CEBPA | 17094 | -2.578 | -0.2823 | Yes | ||
| 126 | ASXL2 | 17137 | -2.617 | -0.2743 | Yes | ||
| 127 | ETF1 | 17168 | -2.649 | -0.2656 | Yes | ||
| 128 | UBR1 | 17516 | -2.978 | -0.2727 | Yes | ||
| 129 | APRIN | 17589 | -3.058 | -0.2647 | Yes | ||
| 130 | RAP2C | 17595 | -3.061 | -0.2530 | Yes | ||
| 131 | ZMYND11 | 17651 | -3.111 | -0.2438 | Yes | ||
| 132 | HBP1 | 17690 | -3.155 | -0.2336 | Yes | ||
| 133 | INSIG1 | 17857 | -3.367 | -0.2294 | Yes | ||
| 134 | HIPK1 | 17930 | -3.486 | -0.2197 | Yes | ||
| 135 | SLMAP | 17942 | -3.502 | -0.2066 | Yes | ||
| 136 | EIF4G2 | 17956 | -3.525 | -0.1935 | Yes | ||
| 137 | KHDRBS1 | 17970 | -3.552 | -0.1804 | Yes | ||
| 138 | SLC38A2 | 17971 | -3.552 | -0.1665 | Yes | ||
| 139 | CNOT7 | 18059 | -3.681 | -0.1568 | Yes | ||
| 140 | ROD1 | 18083 | -3.714 | -0.1435 | Yes | ||
| 141 | CXCR4 | 18137 | -3.818 | -0.1315 | Yes | ||
| 142 | BCL11B | 18292 | -4.156 | -0.1236 | Yes | ||
| 143 | CHD4 | 18302 | -4.175 | -0.1078 | Yes | ||
| 144 | EIF4G3 | 18405 | -4.479 | -0.0958 | Yes | ||
| 145 | BRD7 | 18536 | -5.133 | -0.0828 | Yes | ||
| 146 | SATB1 | 18546 | -5.244 | -0.0628 | Yes | ||
| 147 | ZFYVE20 | 18565 | -5.476 | -0.0424 | Yes | ||
| 148 | GTF2I | 18569 | -5.525 | -0.0209 | Yes | ||
| 149 | APPBP2 | 18595 | -5.998 | 0.0011 | Yes |
