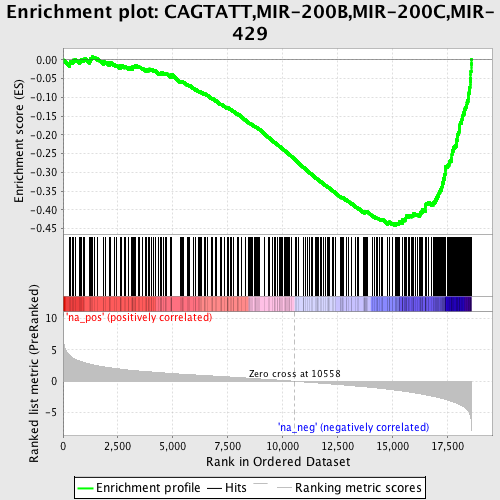
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | CAGTATT,MIR-200B,MIR-200C,MIR-429 |
| Enrichment Score (ES) | -0.4433695 |
| Normalized Enrichment Score (NES) | -2.3074133 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PPM1F | 307 | 4.090 | -0.0100 | No | ||
| 2 | TRIM2 | 320 | 4.052 | -0.0039 | No | ||
| 3 | PCAF | 429 | 3.745 | -0.0036 | No | ||
| 4 | HRB | 478 | 3.665 | -0.0001 | No | ||
| 5 | DLC1 | 562 | 3.505 | 0.0012 | No | ||
| 6 | EIF2B5 | 751 | 3.212 | -0.0038 | No | ||
| 7 | ETV5 | 791 | 3.153 | -0.0007 | No | ||
| 8 | IGSF3 | 843 | 3.095 | 0.0017 | No | ||
| 9 | SMARCD1 | 943 | 2.995 | 0.0012 | No | ||
| 10 | IHPK1 | 993 | 2.942 | 0.0034 | No | ||
| 11 | BAT3 | 1212 | 2.739 | -0.0039 | No | ||
| 12 | PTHLH | 1236 | 2.723 | -0.0007 | No | ||
| 13 | PPP1R10 | 1237 | 2.722 | 0.0039 | No | ||
| 14 | RKHD2 | 1302 | 2.675 | 0.0048 | No | ||
| 15 | SRF | 1321 | 2.658 | 0.0082 | No | ||
| 16 | NDST1 | 1423 | 2.579 | 0.0070 | No | ||
| 17 | RPS6KB1 | 1566 | 2.477 | 0.0034 | No | ||
| 18 | EGR3 | 1848 | 2.302 | -0.0082 | No | ||
| 19 | CREB5 | 1852 | 2.299 | -0.0045 | No | ||
| 20 | PPP1R9B | 1951 | 2.244 | -0.0062 | No | ||
| 21 | CNTFR | 2102 | 2.166 | -0.0108 | No | ||
| 22 | MMD2 | 2106 | 2.163 | -0.0073 | No | ||
| 23 | MTUS1 | 2149 | 2.144 | -0.0061 | No | ||
| 24 | RDH10 | 2325 | 2.060 | -0.0122 | No | ||
| 25 | RPS6KA2 | 2437 | 2.017 | -0.0149 | No | ||
| 26 | MXD3 | 2603 | 1.943 | -0.0207 | No | ||
| 27 | COPS8 | 2606 | 1.942 | -0.0176 | No | ||
| 28 | MGAT2 | 2612 | 1.940 | -0.0147 | No | ||
| 29 | PPP4R2 | 2676 | 1.914 | -0.0149 | No | ||
| 30 | ACE2 | 2798 | 1.866 | -0.0184 | No | ||
| 31 | DNMT3A | 2847 | 1.842 | -0.0180 | No | ||
| 32 | AP1S2 | 2981 | 1.787 | -0.0223 | No | ||
| 33 | KLF12 | 2985 | 1.785 | -0.0195 | No | ||
| 34 | SLC16A2 | 3110 | 1.747 | -0.0234 | No | ||
| 35 | FEZ2 | 3162 | 1.732 | -0.0233 | No | ||
| 36 | HECTD2 | 3174 | 1.728 | -0.0210 | No | ||
| 37 | PHACTR3 | 3179 | 1.727 | -0.0184 | No | ||
| 38 | SBF1 | 3197 | 1.723 | -0.0164 | No | ||
| 39 | FOXK1 | 3270 | 1.699 | -0.0176 | No | ||
| 40 | ARHGDIA | 3281 | 1.696 | -0.0153 | No | ||
| 41 | BRWD1 | 3316 | 1.684 | -0.0143 | No | ||
| 42 | RKHD3 | 3438 | 1.647 | -0.0182 | No | ||
| 43 | SCN3B | 3499 | 1.627 | -0.0188 | No | ||
| 44 | LYPLA2 | 3625 | 1.589 | -0.0230 | No | ||
| 45 | TUBB3 | 3750 | 1.552 | -0.0272 | No | ||
| 46 | PAK7 | 3801 | 1.536 | -0.0274 | No | ||
| 47 | LRP1B | 3810 | 1.534 | -0.0253 | No | ||
| 48 | GPR146 | 3891 | 1.513 | -0.0271 | No | ||
| 49 | NPC1 | 3894 | 1.512 | -0.0247 | No | ||
| 50 | RNF5 | 3930 | 1.505 | -0.0241 | No | ||
| 51 | ELAVL2 | 4014 | 1.483 | -0.0262 | No | ||
| 52 | GLI3 | 4028 | 1.480 | -0.0245 | No | ||
| 53 | ETS1 | 4141 | 1.449 | -0.0282 | No | ||
| 54 | NRP2 | 4193 | 1.437 | -0.0286 | No | ||
| 55 | FGFR2 | 4367 | 1.390 | -0.0357 | No | ||
| 56 | CDH11 | 4428 | 1.375 | -0.0367 | No | ||
| 57 | BASP1 | 4437 | 1.373 | -0.0349 | No | ||
| 58 | PRKAR2B | 4477 | 1.362 | -0.0348 | No | ||
| 59 | CSMD3 | 4488 | 1.359 | -0.0331 | No | ||
| 60 | NFIA | 4585 | 1.334 | -0.0361 | No | ||
| 61 | SLC14A1 | 4652 | 1.319 | -0.0375 | No | ||
| 62 | LIN7B | 4665 | 1.317 | -0.0360 | No | ||
| 63 | PCDH8 | 4732 | 1.302 | -0.0374 | No | ||
| 64 | HS3ST1 | 4893 | 1.261 | -0.0441 | No | ||
| 65 | LCP1 | 4934 | 1.252 | -0.0442 | No | ||
| 66 | KCND2 | 4938 | 1.252 | -0.0423 | No | ||
| 67 | ADCY2 | 4940 | 1.251 | -0.0402 | No | ||
| 68 | ATRX | 4951 | 1.248 | -0.0387 | No | ||
| 69 | PIN1 | 5339 | 1.154 | -0.0580 | No | ||
| 70 | FMR1 | 5381 | 1.144 | -0.0583 | No | ||
| 71 | SOX2 | 5440 | 1.128 | -0.0596 | No | ||
| 72 | FBXW2 | 5447 | 1.126 | -0.0581 | No | ||
| 73 | FGD1 | 5502 | 1.115 | -0.0592 | No | ||
| 74 | PCTK1 | 5667 | 1.084 | -0.0663 | No | ||
| 75 | NR5A2 | 5712 | 1.072 | -0.0670 | No | ||
| 76 | CACNA1C | 5771 | 1.060 | -0.0684 | No | ||
| 77 | SOX1 | 5923 | 1.032 | -0.0749 | No | ||
| 78 | SCD | 6036 | 1.008 | -0.0794 | No | ||
| 79 | SLC1A2 | 6048 | 1.005 | -0.0783 | No | ||
| 80 | PLK2 | 6174 | 0.977 | -0.0835 | No | ||
| 81 | LRRTM3 | 6224 | 0.968 | -0.0846 | No | ||
| 82 | NDN | 6260 | 0.961 | -0.0849 | No | ||
| 83 | CASR | 6327 | 0.945 | -0.0870 | No | ||
| 84 | PPP2R5C | 6440 | 0.921 | -0.0916 | No | ||
| 85 | PTPN13 | 6456 | 0.918 | -0.0908 | No | ||
| 86 | RHOA | 6482 | 0.912 | -0.0907 | No | ||
| 87 | EIF5B | 6566 | 0.897 | -0.0937 | No | ||
| 88 | AMOTL2 | 6751 | 0.862 | -0.1024 | No | ||
| 89 | ACVR2A | 6808 | 0.850 | -0.1040 | No | ||
| 90 | TUBB2A | 6818 | 0.847 | -0.1031 | No | ||
| 91 | NAB1 | 6947 | 0.821 | -0.1088 | No | ||
| 92 | GATA4 | 6980 | 0.815 | -0.1092 | No | ||
| 93 | NXPH1 | 7174 | 0.777 | -0.1184 | No | ||
| 94 | MPP5 | 7209 | 0.770 | -0.1190 | No | ||
| 95 | NAP1L5 | 7230 | 0.766 | -0.1188 | No | ||
| 96 | BHLHB3 | 7339 | 0.741 | -0.1235 | No | ||
| 97 | HMGB3 | 7473 | 0.713 | -0.1296 | No | ||
| 98 | PLCL1 | 7477 | 0.712 | -0.1286 | No | ||
| 99 | SDC2 | 7491 | 0.710 | -0.1281 | No | ||
| 100 | HLF | 7493 | 0.709 | -0.1270 | No | ||
| 101 | GIT2 | 7533 | 0.699 | -0.1280 | No | ||
| 102 | KCNK2 | 7638 | 0.677 | -0.1325 | No | ||
| 103 | LHFP | 7686 | 0.669 | -0.1340 | No | ||
| 104 | HDHD2 | 7759 | 0.655 | -0.1368 | No | ||
| 105 | GPM6A | 7774 | 0.651 | -0.1365 | No | ||
| 106 | TCF2 | 7929 | 0.616 | -0.1439 | No | ||
| 107 | PKIA | 7960 | 0.611 | -0.1445 | No | ||
| 108 | NEGR1 | 8013 | 0.598 | -0.1464 | No | ||
| 109 | TNFRSF11B | 8116 | 0.574 | -0.1510 | No | ||
| 110 | SLC38A4 | 8294 | 0.537 | -0.1598 | No | ||
| 111 | SEMA6D | 8429 | 0.506 | -0.1663 | No | ||
| 112 | DACH1 | 8491 | 0.494 | -0.1688 | No | ||
| 113 | TAF4 | 8513 | 0.489 | -0.1692 | No | ||
| 114 | USP25 | 8553 | 0.477 | -0.1705 | No | ||
| 115 | PTBP1 | 8605 | 0.467 | -0.1725 | No | ||
| 116 | ATXN1 | 8633 | 0.460 | -0.1732 | No | ||
| 117 | MFAP5 | 8710 | 0.443 | -0.1766 | No | ||
| 118 | TRAM1L1 | 8752 | 0.431 | -0.1782 | No | ||
| 119 | LEPR | 8766 | 0.427 | -0.1782 | No | ||
| 120 | AKAP2 | 8825 | 0.412 | -0.1807 | No | ||
| 121 | CORO1C | 8878 | 0.399 | -0.1828 | No | ||
| 122 | SYT1 | 8896 | 0.395 | -0.1831 | No | ||
| 123 | ARHGEF17 | 8951 | 0.383 | -0.1854 | No | ||
| 124 | GNAI3 | 9183 | 0.326 | -0.1975 | No | ||
| 125 | CBX4 | 9364 | 0.289 | -0.2069 | No | ||
| 126 | PRDM1 | 9408 | 0.281 | -0.2088 | No | ||
| 127 | PHF21A | 9550 | 0.250 | -0.2161 | No | ||
| 128 | MYT1 | 9627 | 0.228 | -0.2199 | No | ||
| 129 | EPS8 | 9639 | 0.225 | -0.2201 | No | ||
| 130 | ACY1L2 | 9664 | 0.219 | -0.2210 | No | ||
| 131 | AFF3 | 9773 | 0.198 | -0.2266 | No | ||
| 132 | KCNQ4 | 9779 | 0.196 | -0.2266 | No | ||
| 133 | B3GNT1 | 9858 | 0.178 | -0.2305 | No | ||
| 134 | HIC2 | 9872 | 0.172 | -0.2310 | No | ||
| 135 | FBXW11 | 9893 | 0.167 | -0.2318 | No | ||
| 136 | CAB39 | 9966 | 0.150 | -0.2355 | No | ||
| 137 | RGL1 | 9993 | 0.144 | -0.2367 | No | ||
| 138 | PARD6B | 10112 | 0.116 | -0.2429 | No | ||
| 139 | NUP153 | 10151 | 0.107 | -0.2448 | No | ||
| 140 | DUSP1 | 10152 | 0.107 | -0.2446 | No | ||
| 141 | GLRA2 | 10166 | 0.101 | -0.2452 | No | ||
| 142 | CCNJ | 10225 | 0.085 | -0.2482 | No | ||
| 143 | PLS3 | 10249 | 0.078 | -0.2494 | No | ||
| 144 | ZFPM2 | 10318 | 0.059 | -0.2530 | No | ||
| 145 | KCTD8 | 10333 | 0.055 | -0.2536 | No | ||
| 146 | ST6GALNAC5 | 10394 | 0.038 | -0.2569 | No | ||
| 147 | KLF10 | 10612 | -0.013 | -0.2687 | No | ||
| 148 | PCTK2 | 10632 | -0.019 | -0.2697 | No | ||
| 149 | SEC23A | 10722 | -0.042 | -0.2745 | No | ||
| 150 | EFNA1 | 10939 | -0.096 | -0.2862 | No | ||
| 151 | DNAJB5 | 10940 | -0.096 | -0.2860 | No | ||
| 152 | PLEKHC1 | 10957 | -0.100 | -0.2867 | No | ||
| 153 | PKD1 | 11036 | -0.122 | -0.2908 | No | ||
| 154 | EFNB2 | 11152 | -0.149 | -0.2968 | No | ||
| 155 | BAP1 | 11249 | -0.176 | -0.3018 | No | ||
| 156 | EDG2 | 11303 | -0.188 | -0.3044 | No | ||
| 157 | RAB21 | 11343 | -0.198 | -0.3062 | No | ||
| 158 | LRP1 | 11519 | -0.238 | -0.3154 | No | ||
| 159 | FHOD1 | 11556 | -0.248 | -0.3169 | No | ||
| 160 | YPEL2 | 11613 | -0.263 | -0.3196 | No | ||
| 161 | UBE2B | 11616 | -0.263 | -0.3192 | No | ||
| 162 | CLASP2 | 11635 | -0.268 | -0.3198 | No | ||
| 163 | NPTX1 | 11752 | -0.300 | -0.3256 | No | ||
| 164 | FHL1 | 11794 | -0.315 | -0.3273 | No | ||
| 165 | CRH | 11889 | -0.340 | -0.3319 | No | ||
| 166 | PAM | 11939 | -0.351 | -0.3340 | No | ||
| 167 | IER5 | 12028 | -0.377 | -0.3382 | No | ||
| 168 | GATA2 | 12074 | -0.390 | -0.3400 | No | ||
| 169 | SNAP25 | 12104 | -0.397 | -0.3410 | No | ||
| 170 | EIF2S1 | 12150 | -0.408 | -0.3427 | No | ||
| 171 | RND3 | 12264 | -0.439 | -0.3482 | No | ||
| 172 | SCHIP1 | 12337 | -0.458 | -0.3514 | No | ||
| 173 | STX1A | 12403 | -0.476 | -0.3541 | No | ||
| 174 | TEAD1 | 12634 | -0.542 | -0.3658 | No | ||
| 175 | DGKA | 12649 | -0.546 | -0.3657 | No | ||
| 176 | KCTD15 | 12706 | -0.566 | -0.3678 | No | ||
| 177 | SSH2 | 12718 | -0.569 | -0.3675 | No | ||
| 178 | SYVN1 | 12734 | -0.573 | -0.3673 | No | ||
| 179 | DAG1 | 12758 | -0.579 | -0.3676 | No | ||
| 180 | NIPA1 | 12893 | -0.615 | -0.3739 | No | ||
| 181 | PAPOLA | 12900 | -0.615 | -0.3732 | No | ||
| 182 | GEM | 13003 | -0.642 | -0.3777 | No | ||
| 183 | ESRRG | 13159 | -0.691 | -0.3851 | No | ||
| 184 | THAP1 | 13160 | -0.691 | -0.3839 | No | ||
| 185 | TMEFF2 | 13336 | -0.751 | -0.3923 | No | ||
| 186 | TARDBP | 13395 | -0.768 | -0.3942 | No | ||
| 187 | OCLN | 13474 | -0.790 | -0.3971 | No | ||
| 188 | NPM1 | 13668 | -0.854 | -0.4063 | No | ||
| 189 | DDX3Y | 13721 | -0.868 | -0.4077 | No | ||
| 190 | NGEF | 13723 | -0.869 | -0.4063 | No | ||
| 191 | TUBB | 13724 | -0.869 | -0.4048 | No | ||
| 192 | SULF1 | 13767 | -0.885 | -0.4056 | No | ||
| 193 | UBE2D1 | 13769 | -0.885 | -0.4042 | No | ||
| 194 | PALM2 | 13823 | -0.897 | -0.4056 | No | ||
| 195 | AMFR | 13831 | -0.901 | -0.4045 | No | ||
| 196 | UBE2I | 13886 | -0.920 | -0.4059 | No | ||
| 197 | DCUN1D1 | 14112 | -0.998 | -0.4166 | No | ||
| 198 | ORMDL3 | 14177 | -1.021 | -0.4184 | No | ||
| 199 | PDCD10 | 14274 | -1.054 | -0.4219 | No | ||
| 200 | KCNJ2 | 14314 | -1.069 | -0.4223 | No | ||
| 201 | QKI | 14346 | -1.078 | -0.4222 | No | ||
| 202 | GDI2 | 14437 | -1.115 | -0.4252 | No | ||
| 203 | NOG | 14503 | -1.136 | -0.4269 | No | ||
| 204 | GOSR2 | 14548 | -1.157 | -0.4274 | No | ||
| 205 | PCNP | 14554 | -1.160 | -0.4257 | No | ||
| 206 | CNTN4 | 14792 | -1.245 | -0.4366 | No | ||
| 207 | ARID4B | 14805 | -1.250 | -0.4352 | No | ||
| 208 | YWHAG | 14856 | -1.274 | -0.4358 | No | ||
| 209 | OXR1 | 14861 | -1.275 | -0.4339 | No | ||
| 210 | THRAP1 | 14875 | -1.280 | -0.4325 | No | ||
| 211 | ASF1A | 14999 | -1.333 | -0.4370 | No | ||
| 212 | MAP4K3 | 15024 | -1.348 | -0.4361 | No | ||
| 213 | EGLN1 | 15158 | -1.405 | -0.4410 | Yes | ||
| 214 | CITED2 | 15170 | -1.410 | -0.4393 | Yes | ||
| 215 | RPS6KA3 | 15179 | -1.415 | -0.4374 | Yes | ||
| 216 | HNRPD | 15203 | -1.430 | -0.4363 | Yes | ||
| 217 | ZC3H6 | 15240 | -1.447 | -0.4358 | Yes | ||
| 218 | MAP4K4 | 15300 | -1.473 | -0.4366 | Yes | ||
| 219 | DACT1 | 15319 | -1.480 | -0.4351 | Yes | ||
| 220 | LAMC1 | 15336 | -1.489 | -0.4335 | Yes | ||
| 221 | AEBP2 | 15343 | -1.494 | -0.4314 | Yes | ||
| 222 | EVI5 | 15462 | -1.543 | -0.4353 | Yes | ||
| 223 | ULK2 | 15470 | -1.548 | -0.4331 | Yes | ||
| 224 | PIP5K3 | 15474 | -1.549 | -0.4307 | Yes | ||
| 225 | NRIP1 | 15475 | -1.549 | -0.4281 | Yes | ||
| 226 | HNRPH2 | 15482 | -1.551 | -0.4258 | Yes | ||
| 227 | CHD9 | 15562 | -1.591 | -0.4275 | Yes | ||
| 228 | OLIG3 | 15579 | -1.599 | -0.4257 | Yes | ||
| 229 | MATR3 | 15617 | -1.615 | -0.4251 | Yes | ||
| 230 | GOLGA7 | 15620 | -1.616 | -0.4225 | Yes | ||
| 231 | DNAJB6 | 15628 | -1.620 | -0.4202 | Yes | ||
| 232 | CLIC4 | 15634 | -1.624 | -0.4177 | Yes | ||
| 233 | CSNK1G3 | 15661 | -1.636 | -0.4165 | Yes | ||
| 234 | RABIF | 15738 | -1.676 | -0.4178 | Yes | ||
| 235 | SMURF2 | 15755 | -1.683 | -0.4159 | Yes | ||
| 236 | DMRT2 | 15806 | -1.712 | -0.4158 | Yes | ||
| 237 | NTF3 | 15875 | -1.751 | -0.4166 | Yes | ||
| 238 | PSIP1 | 15927 | -1.781 | -0.4164 | Yes | ||
| 239 | FN1 | 15948 | -1.796 | -0.4145 | Yes | ||
| 240 | SLC6A1 | 15962 | -1.805 | -0.4122 | Yes | ||
| 241 | FLI1 | 15984 | -1.817 | -0.4104 | Yes | ||
| 242 | PPAP2B | 16073 | -1.868 | -0.4121 | Yes | ||
| 243 | RAC1 | 16159 | -1.919 | -0.4135 | Yes | ||
| 244 | ELF2 | 16259 | -1.974 | -0.4157 | Yes | ||
| 245 | NCOA2 | 16268 | -1.980 | -0.4128 | Yes | ||
| 246 | PCDH7 | 16278 | -1.986 | -0.4100 | Yes | ||
| 247 | TSC22D1 | 16285 | -1.992 | -0.4070 | Yes | ||
| 248 | KLF4 | 16323 | -2.019 | -0.4057 | Yes | ||
| 249 | FBXO33 | 16368 | -2.054 | -0.4047 | Yes | ||
| 250 | OGT | 16374 | -2.058 | -0.4015 | Yes | ||
| 251 | MAFG | 16386 | -2.065 | -0.3987 | Yes | ||
| 252 | WDFY3 | 16508 | -2.150 | -0.4018 | Yes | ||
| 253 | YTHDF3 | 16512 | -2.156 | -0.3983 | Yes | ||
| 254 | PPM1B | 16513 | -2.157 | -0.3947 | Yes | ||
| 255 | ZIC3 | 16516 | -2.160 | -0.3913 | Yes | ||
| 256 | PHF6 | 16528 | -2.166 | -0.3883 | Yes | ||
| 257 | NR3C1 | 16533 | -2.168 | -0.3849 | Yes | ||
| 258 | E2F3 | 16574 | -2.193 | -0.3834 | Yes | ||
| 259 | CYP1B1 | 16630 | -2.232 | -0.3827 | Yes | ||
| 260 | VLDLR | 16657 | -2.251 | -0.3804 | Yes | ||
| 261 | BCAP29 | 16798 | -2.351 | -0.3842 | Yes | ||
| 262 | SMARCAD1 | 16860 | -2.394 | -0.3835 | Yes | ||
| 263 | NIN | 16882 | -2.410 | -0.3806 | Yes | ||
| 264 | GABPA | 16927 | -2.441 | -0.3790 | Yes | ||
| 265 | PDPK1 | 16973 | -2.477 | -0.3773 | Yes | ||
| 266 | TBP | 16974 | -2.479 | -0.3732 | Yes | ||
| 267 | CDR2 | 17023 | -2.517 | -0.3717 | Yes | ||
| 268 | CNOT4 | 17025 | -2.520 | -0.3675 | Yes | ||
| 269 | BIRC4 | 17063 | -2.550 | -0.3653 | Yes | ||
| 270 | PHTF2 | 17081 | -2.567 | -0.3620 | Yes | ||
| 271 | MSN | 17115 | -2.593 | -0.3595 | Yes | ||
| 272 | MARCKS | 17128 | -2.603 | -0.3558 | Yes | ||
| 273 | LASS6 | 17140 | -2.625 | -0.3520 | Yes | ||
| 274 | MMD | 17179 | -2.659 | -0.3497 | Yes | ||
| 275 | LRP4 | 17184 | -2.667 | -0.3455 | Yes | ||
| 276 | SEMA3F | 17245 | -2.723 | -0.3442 | Yes | ||
| 277 | TSGA14 | 17247 | -2.724 | -0.3398 | Yes | ||
| 278 | CLASP1 | 17269 | -2.744 | -0.3363 | Yes | ||
| 279 | ANK3 | 17287 | -2.765 | -0.3327 | Yes | ||
| 280 | AKAP7 | 17290 | -2.767 | -0.3282 | Yes | ||
| 281 | TRIM33 | 17323 | -2.795 | -0.3253 | Yes | ||
| 282 | FBXW7 | 17337 | -2.806 | -0.3213 | Yes | ||
| 283 | PUM2 | 17343 | -2.810 | -0.3169 | Yes | ||
| 284 | ADIPOR2 | 17380 | -2.838 | -0.3142 | Yes | ||
| 285 | RANBP10 | 17386 | -2.847 | -0.3097 | Yes | ||
| 286 | MYB | 17394 | -2.858 | -0.3054 | Yes | ||
| 287 | RECK | 17413 | -2.876 | -0.3016 | Yes | ||
| 288 | NEDD1 | 17414 | -2.878 | -0.2968 | Yes | ||
| 289 | PARP6 | 17416 | -2.878 | -0.2920 | Yes | ||
| 290 | HNRPU | 17440 | -2.897 | -0.2885 | Yes | ||
| 291 | NBR1 | 17443 | -2.899 | -0.2838 | Yes | ||
| 292 | SESN1 | 17517 | -2.980 | -0.2828 | Yes | ||
| 293 | RTF1 | 17546 | -3.013 | -0.2793 | Yes | ||
| 294 | APRIN | 17589 | -3.058 | -0.2765 | Yes | ||
| 295 | RAP2C | 17595 | -3.061 | -0.2717 | Yes | ||
| 296 | TOB1 | 17634 | -3.095 | -0.2687 | Yes | ||
| 297 | SERINC1 | 17696 | -3.165 | -0.2667 | Yes | ||
| 298 | NFYA | 17710 | -3.180 | -0.2622 | Yes | ||
| 299 | SLC23A2 | 17721 | -3.193 | -0.2574 | Yes | ||
| 300 | TIAL1 | 17722 | -3.194 | -0.2521 | Yes | ||
| 301 | ANLN | 17729 | -3.202 | -0.2471 | Yes | ||
| 302 | TCEB1 | 17730 | -3.203 | -0.2418 | Yes | ||
| 303 | DDX3X | 17776 | -3.265 | -0.2388 | Yes | ||
| 304 | SFRS2 | 17780 | -3.269 | -0.2335 | Yes | ||
| 305 | KLF9 | 17841 | -3.344 | -0.2312 | Yes | ||
| 306 | LFNG | 17896 | -3.426 | -0.2285 | Yes | ||
| 307 | HIPK1 | 17930 | -3.486 | -0.2245 | Yes | ||
| 308 | CRKL | 17939 | -3.501 | -0.2191 | Yes | ||
| 309 | OSBPL11 | 17943 | -3.502 | -0.2135 | Yes | ||
| 310 | KHDRBS1 | 17970 | -3.552 | -0.2090 | Yes | ||
| 311 | SLC38A2 | 17971 | -3.552 | -0.2031 | Yes | ||
| 312 | CNN3 | 17986 | -3.581 | -0.1979 | Yes | ||
| 313 | KRAS | 18026 | -3.637 | -0.1940 | Yes | ||
| 314 | RANBP9 | 18046 | -3.662 | -0.1889 | Yes | ||
| 315 | CNOT7 | 18059 | -3.681 | -0.1835 | Yes | ||
| 316 | RAP1B | 18070 | -3.696 | -0.1779 | Yes | ||
| 317 | TBK1 | 18086 | -3.732 | -0.1725 | Yes | ||
| 318 | YWHAB | 18089 | -3.736 | -0.1664 | Yes | ||
| 319 | PPP2CA | 18149 | -3.839 | -0.1632 | Yes | ||
| 320 | ARL8B | 18176 | -3.891 | -0.1582 | Yes | ||
| 321 | NUDT4 | 18190 | -3.911 | -0.1524 | Yes | ||
| 322 | HNRPK | 18220 | -3.977 | -0.1474 | Yes | ||
| 323 | ACACA | 18233 | -4.013 | -0.1413 | Yes | ||
| 324 | BCL11B | 18292 | -4.156 | -0.1376 | Yes | ||
| 325 | ATP2A2 | 18299 | -4.171 | -0.1310 | Yes | ||
| 326 | RAB6IP1 | 18343 | -4.274 | -0.1262 | Yes | ||
| 327 | PRKACB | 18370 | -4.348 | -0.1204 | Yes | ||
| 328 | SFRS1 | 18407 | -4.486 | -0.1150 | Yes | ||
| 329 | RHOT1 | 18415 | -4.531 | -0.1078 | Yes | ||
| 330 | ARID2 | 18457 | -4.666 | -0.1023 | Yes | ||
| 331 | BRMS1L | 18487 | -4.772 | -0.0959 | Yes | ||
| 332 | CFL2 | 18494 | -4.838 | -0.0882 | Yes | ||
| 333 | DDEF1 | 18510 | -4.920 | -0.0809 | Yes | ||
| 334 | HS2ST1 | 18529 | -5.048 | -0.0735 | Yes | ||
| 335 | ITPR1 | 18547 | -5.257 | -0.0656 | Yes | ||
| 336 | ICK | 18550 | -5.291 | -0.0570 | Yes | ||
| 337 | ZFYVE20 | 18565 | -5.476 | -0.0486 | Yes | ||
| 338 | CALU | 18582 | -5.802 | -0.0398 | Yes | ||
| 339 | SLC6A6 | 18586 | -5.883 | -0.0302 | Yes | ||
| 340 | CDYL | 18596 | -6.035 | -0.0207 | Yes | ||
| 341 | UBQLN1 | 18597 | -6.067 | -0.0106 | Yes | ||
| 342 | FREQ | 18608 | -6.962 | 0.0004 | Yes |
