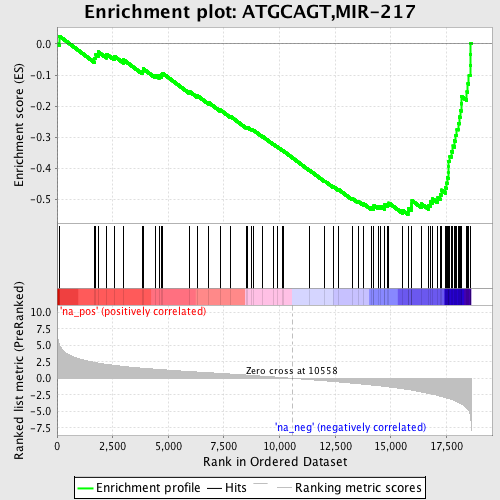
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ATGCAGT,MIR-217 |
| Enrichment Score (ES) | -0.5476783 |
| Normalized Enrichment Score (NES) | -2.3224714 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CAPN3 | 106 | 5.012 | 0.0249 | No | ||
| 2 | PSMF1 | 1691 | 2.396 | -0.0459 | No | ||
| 3 | KCNA3 | 1740 | 2.365 | -0.0340 | No | ||
| 4 | HIVEP3 | 1846 | 2.305 | -0.0255 | No | ||
| 5 | TACC2 | 2217 | 2.117 | -0.0325 | No | ||
| 6 | NEUROD1 | 2572 | 1.961 | -0.0397 | No | ||
| 7 | KLF12 | 2985 | 1.785 | -0.0510 | No | ||
| 8 | ST18 | 3831 | 1.529 | -0.0872 | No | ||
| 9 | STAT5B | 3867 | 1.521 | -0.0798 | No | ||
| 10 | GRIK2 | 4404 | 1.381 | -0.1002 | No | ||
| 11 | NFIA | 4585 | 1.334 | -0.1018 | No | ||
| 12 | BCL11A | 4692 | 1.312 | -0.0995 | No | ||
| 13 | CHN2 | 4736 | 1.301 | -0.0938 | No | ||
| 14 | SEPT4 | 5931 | 1.030 | -0.1520 | No | ||
| 15 | PAQR3 | 6297 | 0.952 | -0.1658 | No | ||
| 16 | ACVR2A | 6808 | 0.850 | -0.1881 | No | ||
| 17 | A2BP1 | 7334 | 0.742 | -0.2119 | No | ||
| 18 | GPM6A | 7774 | 0.651 | -0.2316 | No | ||
| 19 | DACH1 | 8491 | 0.494 | -0.2672 | No | ||
| 20 | DLGAP2 | 8550 | 0.478 | -0.2674 | No | ||
| 21 | SLITRK3 | 8724 | 0.438 | -0.2741 | No | ||
| 22 | AKAP2 | 8825 | 0.412 | -0.2770 | No | ||
| 23 | TBC1D15 | 9239 | 0.314 | -0.2973 | No | ||
| 24 | FBN2 | 9733 | 0.206 | -0.3226 | No | ||
| 25 | FBXW11 | 9893 | 0.167 | -0.3302 | No | ||
| 26 | POLG | 10111 | 0.116 | -0.3412 | No | ||
| 27 | ADAMTS5 | 10172 | 0.100 | -0.3438 | No | ||
| 28 | CUL5 | 11363 | -0.202 | -0.4068 | No | ||
| 29 | MAMDC2 | 12030 | -0.377 | -0.4404 | No | ||
| 30 | STX1A | 12403 | -0.476 | -0.4575 | No | ||
| 31 | ELOVL4 | 12668 | -0.551 | -0.4684 | No | ||
| 32 | ZCCHC2 | 13268 | -0.728 | -0.4963 | No | ||
| 33 | SLC39A10 | 13527 | -0.807 | -0.5053 | No | ||
| 34 | STAG2 | 13758 | -0.881 | -0.5123 | No | ||
| 35 | DYRK1A | 14124 | -1.003 | -0.5258 | No | ||
| 36 | BAI3 | 14220 | -1.034 | -0.5246 | No | ||
| 37 | JOSD3 | 14238 | -1.038 | -0.5192 | No | ||
| 38 | GDI2 | 14437 | -1.115 | -0.5231 | No | ||
| 39 | ATP11C | 14527 | -1.145 | -0.5209 | No | ||
| 40 | TMSB4X | 14725 | -1.219 | -0.5240 | No | ||
| 41 | STRBP | 14730 | -1.220 | -0.5168 | No | ||
| 42 | YWHAG | 14856 | -1.274 | -0.5157 | No | ||
| 43 | SOX11 | 14912 | -1.297 | -0.5108 | No | ||
| 44 | NR4A2 | 15532 | -1.575 | -0.5345 | No | ||
| 45 | EZH2 | 15777 | -1.693 | -0.5373 | Yes | ||
| 46 | DMRT2 | 15806 | -1.712 | -0.5284 | Yes | ||
| 47 | RIN2 | 15916 | -1.775 | -0.5234 | Yes | ||
| 48 | KCTD9 | 15942 | -1.792 | -0.5138 | Yes | ||
| 49 | FN1 | 15948 | -1.796 | -0.5031 | Yes | ||
| 50 | OGT | 16374 | -2.058 | -0.5134 | Yes | ||
| 51 | ATP6V1A | 16707 | -2.279 | -0.5174 | Yes | ||
| 52 | STX6 | 16781 | -2.336 | -0.5070 | Yes | ||
| 53 | HNRPUL1 | 16880 | -2.408 | -0.4976 | Yes | ||
| 54 | MSN | 17115 | -2.593 | -0.4943 | Yes | ||
| 55 | MTPN | 17224 | -2.703 | -0.4836 | Yes | ||
| 56 | SIRT1 | 17264 | -2.740 | -0.4690 | Yes | ||
| 57 | TLK2 | 17454 | -2.905 | -0.4614 | Yes | ||
| 58 | EIF4A2 | 17518 | -2.986 | -0.4465 | Yes | ||
| 59 | RTF1 | 17546 | -3.013 | -0.4296 | Yes | ||
| 60 | APRIN | 17589 | -3.058 | -0.4131 | Yes | ||
| 61 | RAP2C | 17595 | -3.061 | -0.3947 | Yes | ||
| 62 | HNRPA2B1 | 17601 | -3.064 | -0.3762 | Yes | ||
| 63 | ATP1B1 | 17654 | -3.116 | -0.3599 | Yes | ||
| 64 | MAPK1 | 17720 | -3.193 | -0.3439 | Yes | ||
| 65 | SLC31A1 | 17793 | -3.280 | -0.3277 | Yes | ||
| 66 | PCNA | 17864 | -3.379 | -0.3109 | Yes | ||
| 67 | YAF2 | 17919 | -3.470 | -0.2925 | Yes | ||
| 68 | SLC38A2 | 17971 | -3.552 | -0.2736 | Yes | ||
| 69 | KRAS | 18026 | -3.637 | -0.2542 | Yes | ||
| 70 | SENP5 | 18082 | -3.714 | -0.2345 | Yes | ||
| 71 | UBL3 | 18117 | -3.780 | -0.2132 | Yes | ||
| 72 | EHMT1 | 18164 | -3.857 | -0.1921 | Yes | ||
| 73 | MYEF2 | 18183 | -3.898 | -0.1692 | Yes | ||
| 74 | NDEL1 | 18422 | -4.555 | -0.1542 | Yes | ||
| 75 | IVNS1ABP | 18464 | -4.681 | -0.1278 | Yes | ||
| 76 | DYNC1LI2 | 18512 | -4.942 | -0.1001 | Yes | ||
| 77 | ZFYVE20 | 18565 | -5.476 | -0.0694 | Yes | ||
| 78 | PPM1D | 18580 | -5.779 | -0.0348 | Yes | ||
| 79 | APPBP2 | 18595 | -5.998 | 0.0011 | Yes |
