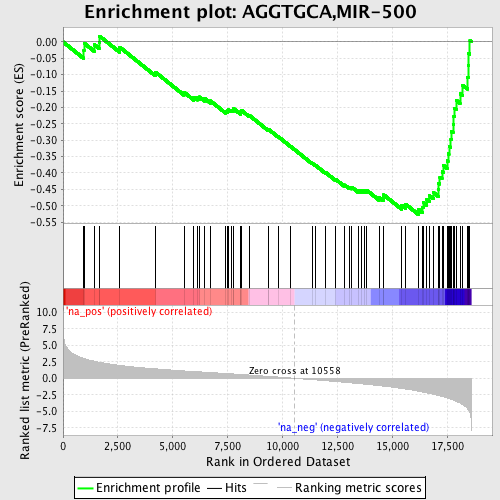
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | AGGTGCA,MIR-500 |
| Enrichment Score (ES) | -0.5267118 |
| Normalized Enrichment Score (NES) | -2.1899064 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.7241144E-4 |
| FWER p-Value | 0.0020 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RTN4RL1 | 946 | 2.991 | -0.0271 | No | ||
| 2 | RNF40 | 958 | 2.982 | -0.0038 | No | ||
| 3 | EGLN2 | 1419 | 2.583 | -0.0080 | No | ||
| 4 | POFUT1 | 1647 | 2.428 | -0.0008 | No | ||
| 5 | SP8 | 1664 | 2.417 | 0.0177 | No | ||
| 6 | CELSR3 | 2574 | 1.960 | -0.0156 | No | ||
| 7 | PPP3CC | 4206 | 1.434 | -0.0921 | No | ||
| 8 | SYT8 | 5510 | 1.113 | -0.1535 | No | ||
| 9 | PHOX2B | 5964 | 1.023 | -0.1697 | No | ||
| 10 | RAI2 | 6108 | 0.990 | -0.1695 | No | ||
| 11 | LRRTM3 | 6224 | 0.968 | -0.1680 | No | ||
| 12 | OSBPL10 | 6462 | 0.918 | -0.1734 | No | ||
| 13 | LBP | 6714 | 0.869 | -0.1800 | No | ||
| 14 | ING4 | 7408 | 0.726 | -0.2115 | No | ||
| 15 | HLF | 7493 | 0.709 | -0.2104 | No | ||
| 16 | HSPA14 | 7528 | 0.700 | -0.2066 | No | ||
| 17 | TSSK2 | 7658 | 0.674 | -0.2082 | No | ||
| 18 | SLC25A26 | 7752 | 0.655 | -0.2080 | No | ||
| 19 | GPM6A | 7774 | 0.651 | -0.2039 | No | ||
| 20 | PLXDC2 | 8094 | 0.580 | -0.2164 | No | ||
| 21 | TBX2 | 8100 | 0.579 | -0.2121 | No | ||
| 22 | CHRD | 8133 | 0.571 | -0.2092 | No | ||
| 23 | HNRPA0 | 8493 | 0.494 | -0.2246 | No | ||
| 24 | GABRB3 | 9366 | 0.288 | -0.2694 | No | ||
| 25 | SHPRH | 9382 | 0.286 | -0.2679 | No | ||
| 26 | BTBD11 | 9812 | 0.188 | -0.2895 | No | ||
| 27 | CIT | 10357 | 0.050 | -0.3184 | No | ||
| 28 | RAB21 | 11343 | -0.198 | -0.3700 | No | ||
| 29 | SPG3A | 11492 | -0.231 | -0.3761 | No | ||
| 30 | NOLA2 | 11958 | -0.356 | -0.3983 | No | ||
| 31 | THAP11 | 12435 | -0.485 | -0.4201 | No | ||
| 32 | PRKCE | 12831 | -0.599 | -0.4366 | No | ||
| 33 | CACNB1 | 13049 | -0.657 | -0.4431 | No | ||
| 34 | ESRRG | 13159 | -0.691 | -0.4434 | No | ||
| 35 | RNF38 | 13473 | -0.790 | -0.4540 | No | ||
| 36 | SEC24C | 13581 | -0.824 | -0.4531 | No | ||
| 37 | DDX3Y | 13721 | -0.868 | -0.4537 | No | ||
| 38 | NFAT5 | 13837 | -0.904 | -0.4527 | No | ||
| 39 | DRD1 | 14402 | -1.102 | -0.4743 | No | ||
| 40 | ABCC4 | 14592 | -1.169 | -0.4751 | No | ||
| 41 | FIGN | 14616 | -1.180 | -0.4669 | No | ||
| 42 | NRF1 | 15433 | -1.530 | -0.4987 | No | ||
| 43 | MATR3 | 15617 | -1.615 | -0.4956 | No | ||
| 44 | UBE2D3 | 16195 | -1.941 | -0.5112 | Yes | ||
| 45 | OGT | 16374 | -2.058 | -0.5043 | Yes | ||
| 46 | PURB | 16416 | -2.082 | -0.4899 | Yes | ||
| 47 | E2F3 | 16574 | -2.193 | -0.4808 | Yes | ||
| 48 | KPNA3 | 16694 | -2.272 | -0.4691 | Yes | ||
| 49 | NCOA1 | 16881 | -2.409 | -0.4598 | Yes | ||
| 50 | PPP4R1 | 17101 | -2.586 | -0.4510 | Yes | ||
| 51 | RBMS1 | 17117 | -2.594 | -0.4310 | Yes | ||
| 52 | SORCS1 | 17172 | -2.652 | -0.4127 | Yes | ||
| 53 | CAMK4 | 17272 | -2.749 | -0.3961 | Yes | ||
| 54 | FBXW7 | 17337 | -2.806 | -0.3771 | Yes | ||
| 55 | MYNN | 17504 | -2.952 | -0.3624 | Yes | ||
| 56 | TIMM8A | 17547 | -3.013 | -0.3406 | Yes | ||
| 57 | APRIN | 17589 | -3.058 | -0.3184 | Yes | ||
| 58 | CREB1 | 17673 | -3.133 | -0.2978 | Yes | ||
| 59 | SOCS2 | 17700 | -3.168 | -0.2738 | Yes | ||
| 60 | DDX3X | 17776 | -3.265 | -0.2518 | Yes | ||
| 61 | PPP2R5E | 17807 | -3.301 | -0.2270 | Yes | ||
| 62 | KLF9 | 17841 | -3.344 | -0.2020 | Yes | ||
| 63 | HIPK1 | 17930 | -3.486 | -0.1789 | Yes | ||
| 64 | DCTN4 | 18094 | -3.748 | -0.1577 | Yes | ||
| 65 | SLC4A7 | 18208 | -3.959 | -0.1321 | Yes | ||
| 66 | KBTBD2 | 18430 | -4.581 | -0.1074 | Yes | ||
| 67 | IVNS1ABP | 18464 | -4.681 | -0.0717 | Yes | ||
| 68 | RAB14 | 18484 | -4.754 | -0.0347 | Yes | ||
| 69 | CUTL1 | 18543 | -5.228 | 0.0039 | Yes |
