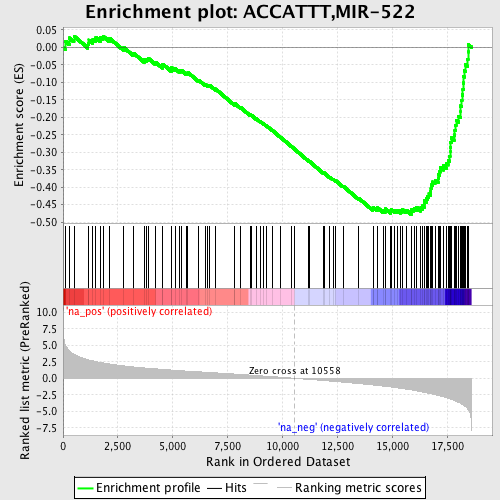
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_truncNotch_versus_absentNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | ACCATTT,MIR-522 |
| Enrichment Score (ES) | -0.47794685 |
| Normalized Enrichment Score (NES) | -2.1410475 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.3413205E-4 |
| FWER p-Value | 0.0030 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CAPN3 | 106 | 5.012 | 0.0173 | No | ||
| 2 | CRIM1 | 282 | 4.175 | 0.0271 | No | ||
| 3 | WDTC1 | 497 | 3.634 | 0.0322 | No | ||
| 4 | FZD2 | 1134 | 2.803 | 0.0107 | No | ||
| 5 | SH3BP5 | 1178 | 2.763 | 0.0211 | No | ||
| 6 | PTPN1 | 1358 | 2.625 | 0.0235 | No | ||
| 7 | UBE2J1 | 1494 | 2.527 | 0.0278 | No | ||
| 8 | CSPG5 | 1694 | 2.395 | 0.0281 | No | ||
| 9 | APH1A | 1820 | 2.323 | 0.0320 | No | ||
| 10 | CNTFR | 2102 | 2.166 | 0.0268 | No | ||
| 11 | SALL1 | 2767 | 1.878 | -0.0004 | No | ||
| 12 | UBE3A | 3207 | 1.719 | -0.0162 | No | ||
| 13 | PTGFRN | 3694 | 1.571 | -0.0353 | No | ||
| 14 | LRP1B | 3810 | 1.534 | -0.0344 | No | ||
| 15 | CTBP1 | 3885 | 1.516 | -0.0315 | No | ||
| 16 | FLRT3 | 4209 | 1.434 | -0.0423 | No | ||
| 17 | PLAG1 | 4525 | 1.351 | -0.0531 | No | ||
| 18 | TNPO2 | 4538 | 1.348 | -0.0476 | No | ||
| 19 | HOXB5 | 4944 | 1.250 | -0.0637 | No | ||
| 20 | ATRX | 4951 | 1.248 | -0.0583 | No | ||
| 21 | WNK3 | 5103 | 1.210 | -0.0609 | No | ||
| 22 | GAB2 | 5315 | 1.159 | -0.0670 | No | ||
| 23 | FMR1 | 5381 | 1.144 | -0.0652 | No | ||
| 24 | PERP | 5626 | 1.091 | -0.0734 | No | ||
| 25 | TCFL5 | 5689 | 1.079 | -0.0718 | No | ||
| 26 | PLK2 | 6174 | 0.977 | -0.0934 | No | ||
| 27 | ARID5B | 6468 | 0.915 | -0.1051 | No | ||
| 28 | SOX9 | 6596 | 0.892 | -0.1078 | No | ||
| 29 | CDS2 | 6671 | 0.877 | -0.1078 | No | ||
| 30 | NAB1 | 6947 | 0.821 | -0.1189 | No | ||
| 31 | RBM22 | 7814 | 0.643 | -0.1627 | No | ||
| 32 | MAFB | 7815 | 0.643 | -0.1598 | No | ||
| 33 | DOCK7 | 8083 | 0.582 | -0.1715 | No | ||
| 34 | PANK2 | 8518 | 0.488 | -0.1927 | No | ||
| 35 | PHYHIPL | 8584 | 0.470 | -0.1941 | No | ||
| 36 | MEF2D | 8804 | 0.420 | -0.2040 | No | ||
| 37 | CBLL1 | 8973 | 0.376 | -0.2113 | No | ||
| 38 | ZFP36L2 | 9133 | 0.335 | -0.2184 | No | ||
| 39 | GPM6B | 9258 | 0.312 | -0.2236 | No | ||
| 40 | RNF2 | 9535 | 0.255 | -0.2374 | No | ||
| 41 | NPAT | 9921 | 0.159 | -0.2574 | No | ||
| 42 | LRCH2 | 10419 | 0.032 | -0.2842 | No | ||
| 43 | SP4 | 10550 | 0.002 | -0.2912 | No | ||
| 44 | TBC1D8 | 11192 | -0.160 | -0.3251 | No | ||
| 45 | TLX1 | 11228 | -0.168 | -0.3262 | No | ||
| 46 | ARHGAP5 | 11872 | -0.335 | -0.3594 | No | ||
| 47 | AQP9 | 11894 | -0.341 | -0.3590 | No | ||
| 48 | TRPM7 | 12160 | -0.410 | -0.3714 | No | ||
| 49 | UBE2L3 | 12302 | -0.447 | -0.3770 | No | ||
| 50 | JUN | 12417 | -0.481 | -0.3809 | No | ||
| 51 | DAG1 | 12758 | -0.579 | -0.3966 | No | ||
| 52 | BHLHB5 | 13472 | -0.789 | -0.4315 | No | ||
| 53 | DYRK1A | 14124 | -1.003 | -0.4621 | No | ||
| 54 | RSBN1 | 14139 | -1.009 | -0.4582 | No | ||
| 55 | NR4A3 | 14316 | -1.070 | -0.4628 | No | ||
| 56 | UBE2N | 14336 | -1.076 | -0.4589 | No | ||
| 57 | TTN | 14598 | -1.171 | -0.4676 | No | ||
| 58 | ING3 | 14674 | -1.204 | -0.4661 | No | ||
| 59 | WDR26 | 14699 | -1.212 | -0.4618 | No | ||
| 60 | ARMCX2 | 14943 | -1.310 | -0.4689 | No | ||
| 61 | PRKG1 | 14960 | -1.315 | -0.4637 | No | ||
| 62 | CASKIN1 | 15118 | -1.385 | -0.4659 | No | ||
| 63 | ETV1 | 15245 | -1.448 | -0.4660 | No | ||
| 64 | NFKBIZ | 15396 | -1.516 | -0.4671 | No | ||
| 65 | YWHAQ | 15469 | -1.548 | -0.4639 | No | ||
| 66 | ZC3H7A | 15638 | -1.625 | -0.4655 | No | ||
| 67 | NFE2L2 | 15869 | -1.748 | -0.4699 | Yes | ||
| 68 | CUL1 | 15887 | -1.757 | -0.4627 | Yes | ||
| 69 | MYH10 | 15992 | -1.822 | -0.4600 | Yes | ||
| 70 | ENC1 | 16103 | -1.886 | -0.4573 | Yes | ||
| 71 | SLC7A6 | 16307 | -2.009 | -0.4590 | Yes | ||
| 72 | AKAP1 | 16378 | -2.059 | -0.4533 | Yes | ||
| 73 | STAG1 | 16457 | -2.109 | -0.4478 | Yes | ||
| 74 | SLC39A9 | 16464 | -2.114 | -0.4384 | Yes | ||
| 75 | CPD | 16549 | -2.177 | -0.4329 | Yes | ||
| 76 | MECP2 | 16612 | -2.221 | -0.4261 | Yes | ||
| 77 | KIF23 | 16675 | -2.259 | -0.4190 | Yes | ||
| 78 | SYNCRIP | 16752 | -2.313 | -0.4125 | Yes | ||
| 79 | CDV3 | 16765 | -2.320 | -0.4025 | Yes | ||
| 80 | PTPN11 | 16795 | -2.348 | -0.3933 | Yes | ||
| 81 | AKAP9 | 16844 | -2.382 | -0.3849 | Yes | ||
| 82 | TOP1 | 16952 | -2.462 | -0.3794 | Yes | ||
| 83 | CEBPA | 17094 | -2.578 | -0.3751 | Yes | ||
| 84 | GNAO1 | 17109 | -2.590 | -0.3640 | Yes | ||
| 85 | CCND2 | 17169 | -2.650 | -0.3550 | Yes | ||
| 86 | PRPF39 | 17199 | -2.683 | -0.3442 | Yes | ||
| 87 | YWHAZ | 17324 | -2.796 | -0.3380 | Yes | ||
| 88 | GOLGA1 | 17481 | -2.932 | -0.3330 | Yes | ||
| 89 | RTF1 | 17546 | -3.013 | -0.3226 | Yes | ||
| 90 | CSNK1A1 | 17614 | -3.081 | -0.3120 | Yes | ||
| 91 | CUGBP2 | 17636 | -3.096 | -0.2989 | Yes | ||
| 92 | GPIAP1 | 17642 | -3.102 | -0.2849 | Yes | ||
| 93 | ATP1B1 | 17654 | -3.116 | -0.2712 | Yes | ||
| 94 | HBP1 | 17690 | -3.155 | -0.2586 | Yes | ||
| 95 | MBNL1 | 17821 | -3.320 | -0.2503 | Yes | ||
| 96 | FEM1C | 17856 | -3.367 | -0.2367 | Yes | ||
| 97 | TOPORS | 17875 | -3.404 | -0.2220 | Yes | ||
| 98 | FOXP1 | 17911 | -3.457 | -0.2080 | Yes | ||
| 99 | KRAS | 18026 | -3.637 | -0.1974 | Yes | ||
| 100 | RBMX | 18107 | -3.763 | -0.1845 | Yes | ||
| 101 | HNRPM | 18113 | -3.772 | -0.1674 | Yes | ||
| 102 | EHMT1 | 18164 | -3.857 | -0.1523 | Yes | ||
| 103 | MYEF2 | 18183 | -3.898 | -0.1354 | Yes | ||
| 104 | HNRPK | 18220 | -3.977 | -0.1191 | Yes | ||
| 105 | KPNB1 | 18242 | -4.033 | -0.1016 | Yes | ||
| 106 | MAN1A2 | 18255 | -4.064 | -0.0836 | Yes | ||
| 107 | ELAVL1 | 18282 | -4.139 | -0.0660 | Yes | ||
| 108 | VAPA | 18335 | -4.256 | -0.0492 | Yes | ||
| 109 | TMEM2 | 18443 | -4.619 | -0.0337 | Yes | ||
| 110 | ARID2 | 18457 | -4.666 | -0.0130 | Yes | ||
| 111 | IVNS1ABP | 18464 | -4.681 | 0.0082 | Yes |
