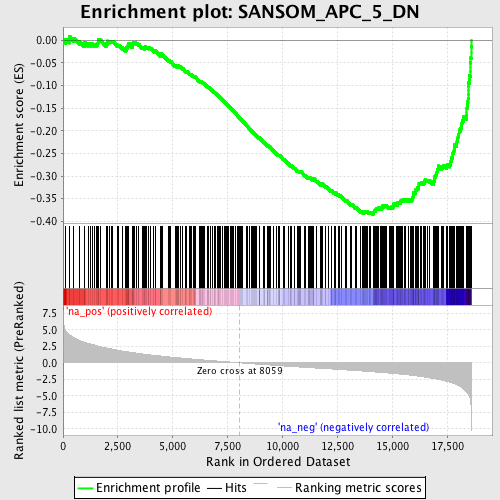
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | SANSOM_APC_5_DN |
| Enrichment Score (ES) | -0.38510856 |
| Normalized Enrichment Score (NES) | -1.9976552 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0272833 |
| FWER p-Value | 0.169 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DDC | 119 | 4.868 | 0.0024 | No | ||
| 2 | SETDB1 | 272 | 4.276 | 0.0019 | No | ||
| 3 | MXI1 | 280 | 4.258 | 0.0094 | No | ||
| 4 | 6330412F12RIK | 492 | 3.799 | 0.0048 | No | ||
| 5 | MAP3K1 | 753 | 3.379 | -0.0033 | No | ||
| 6 | ATP1B1 | 962 | 3.116 | -0.0089 | No | ||
| 7 | TOB1 | 982 | 3.095 | -0.0043 | No | ||
| 8 | ABR | 1136 | 2.931 | -0.0073 | No | ||
| 9 | ADIPOR2 | 1236 | 2.838 | -0.0075 | No | ||
| 10 | ANK3 | 1329 | 2.765 | -0.0075 | No | ||
| 11 | SAR1B | 1435 | 2.660 | -0.0084 | No | ||
| 12 | MTTP | 1517 | 2.584 | -0.0081 | No | ||
| 13 | PTPRC | 1566 | 2.541 | -0.0060 | No | ||
| 14 | CDR2 | 1593 | 2.517 | -0.0028 | No | ||
| 15 | SEC14L1 | 1596 | 2.514 | 0.0017 | No | ||
| 16 | HNRPA1 | 1691 | 2.440 | 0.0010 | No | ||
| 17 | TPH1 | 1958 | 2.252 | -0.0094 | No | ||
| 18 | TRFR2 | 1988 | 2.231 | -0.0069 | No | ||
| 19 | CASP1 | 2022 | 2.206 | -0.0047 | No | ||
| 20 | 5730469M10RIK | 2037 | 2.197 | -0.0014 | No | ||
| 21 | CYFIP2 | 2119 | 2.138 | -0.0020 | No | ||
| 22 | OLFR91 | 2184 | 2.094 | -0.0016 | No | ||
| 23 | CCL5 | 2252 | 2.051 | -0.0015 | No | ||
| 24 | CUL4B | 2475 | 1.908 | -0.0102 | No | ||
| 25 | PAPSS2 | 2546 | 1.867 | -0.0106 | No | ||
| 26 | FAHD1 | 2711 | 1.766 | -0.0163 | No | ||
| 27 | MGST3 | 2852 | 1.686 | -0.0209 | No | ||
| 28 | RORC | 2877 | 1.676 | -0.0191 | No | ||
| 29 | SLC22A18 | 2886 | 1.673 | -0.0165 | No | ||
| 30 | IL18 | 2924 | 1.651 | -0.0155 | No | ||
| 31 | DNASE1 | 2925 | 1.651 | -0.0125 | No | ||
| 32 | CD52 | 2964 | 1.633 | -0.0116 | No | ||
| 33 | CD28 | 2991 | 1.619 | -0.0100 | No | ||
| 34 | JAK2 | 2997 | 1.615 | -0.0073 | No | ||
| 35 | NCF1 | 3152 | 1.546 | -0.0129 | No | ||
| 36 | SRXN1 | 3155 | 1.543 | -0.0102 | No | ||
| 37 | ITGA3 | 3159 | 1.541 | -0.0075 | No | ||
| 38 | KLRA7 | 3194 | 1.525 | -0.0066 | No | ||
| 39 | SIRPA | 3195 | 1.525 | -0.0038 | No | ||
| 40 | ARG2 | 3248 | 1.506 | -0.0039 | No | ||
| 41 | SEC14L2 | 3334 | 1.468 | -0.0059 | No | ||
| 42 | CITED2 | 3446 | 1.410 | -0.0094 | No | ||
| 43 | RGS2 | 3610 | 1.337 | -0.0158 | No | ||
| 44 | CYP3A25 | 3643 | 1.319 | -0.0152 | No | ||
| 45 | ICAM5 | 3724 | 1.286 | -0.0172 | No | ||
| 46 | PAX8 | 3727 | 1.285 | -0.0149 | No | ||
| 47 | MERTK | 3737 | 1.281 | -0.0131 | No | ||
| 48 | CYB5A | 3810 | 1.251 | -0.0147 | No | ||
| 49 | CGGBP1 | 3892 | 1.218 | -0.0169 | No | ||
| 50 | WBP2 | 3897 | 1.217 | -0.0149 | No | ||
| 51 | NUMB | 3975 | 1.190 | -0.0169 | No | ||
| 52 | ERCC5 | 4106 | 1.140 | -0.0220 | No | ||
| 53 | RRAGD | 4195 | 1.109 | -0.0248 | No | ||
| 54 | H2-DMB1 | 4201 | 1.105 | -0.0230 | No | ||
| 55 | SLC7A8 | 4433 | 1.023 | -0.0338 | No | ||
| 56 | LY75 | 4436 | 1.022 | -0.0320 | No | ||
| 57 | SDF4 | 4437 | 1.022 | -0.0301 | No | ||
| 58 | ZDHHC14 | 4467 | 1.013 | -0.0299 | No | ||
| 59 | LTB4R | 4522 | 0.990 | -0.0310 | No | ||
| 60 | TCEA3 | 4784 | 0.902 | -0.0436 | No | ||
| 61 | TYROBP | 4843 | 0.886 | -0.0452 | No | ||
| 62 | ENTPD5 | 4916 | 0.861 | -0.0475 | No | ||
| 63 | CASP3 | 5125 | 0.796 | -0.0575 | No | ||
| 64 | GGT1 | 5155 | 0.787 | -0.0576 | No | ||
| 65 | FAS | 5156 | 0.786 | -0.0562 | No | ||
| 66 | RASSF2 | 5210 | 0.772 | -0.0577 | No | ||
| 67 | IGJ | 5212 | 0.772 | -0.0563 | No | ||
| 68 | CLGN | 5236 | 0.765 | -0.0562 | No | ||
| 69 | CCL25 | 5271 | 0.753 | -0.0566 | No | ||
| 70 | AADAC | 5346 | 0.728 | -0.0593 | No | ||
| 71 | CAPN9 | 5349 | 0.727 | -0.0581 | No | ||
| 72 | ABP1 | 5449 | 0.694 | -0.0623 | No | ||
| 73 | 2310016A09RIK | 5594 | 0.648 | -0.0690 | No | ||
| 74 | GPR12 | 5596 | 0.647 | -0.0678 | No | ||
| 75 | CASP6 | 5640 | 0.635 | -0.0690 | No | ||
| 76 | MYO7A | 5767 | 0.602 | -0.0748 | No | ||
| 77 | CDKN2B | 5782 | 0.599 | -0.0745 | No | ||
| 78 | TGFBI | 5804 | 0.595 | -0.0745 | No | ||
| 79 | 1810046J19RIK | 5868 | 0.576 | -0.0769 | No | ||
| 80 | SLC22A4 | 5924 | 0.562 | -0.0789 | No | ||
| 81 | DGKA | 5967 | 0.546 | -0.0802 | No | ||
| 82 | TRP53INP2 | 6000 | 0.537 | -0.0810 | No | ||
| 83 | MGAT3 | 6015 | 0.532 | -0.0808 | No | ||
| 84 | XDH | 6224 | 0.474 | -0.0913 | No | ||
| 85 | FLT4 | 6227 | 0.473 | -0.0905 | No | ||
| 86 | GZMB | 6276 | 0.459 | -0.0923 | No | ||
| 87 | ADORA2B | 6278 | 0.458 | -0.0915 | No | ||
| 88 | NUDT5 | 6296 | 0.451 | -0.0916 | No | ||
| 89 | TNFRSF17 | 6338 | 0.443 | -0.0931 | No | ||
| 90 | LAPTM5 | 6365 | 0.435 | -0.0937 | No | ||
| 91 | CD48 | 6410 | 0.423 | -0.0953 | No | ||
| 92 | ICAM2 | 6426 | 0.418 | -0.0954 | No | ||
| 93 | EDN3 | 6559 | 0.385 | -0.1019 | No | ||
| 94 | ZNF467 | 6568 | 0.381 | -0.1016 | No | ||
| 95 | BARX2 | 6617 | 0.369 | -0.1036 | No | ||
| 96 | SLC7A9 | 6636 | 0.362 | -0.1039 | No | ||
| 97 | GIMAP4 | 6713 | 0.344 | -0.1074 | No | ||
| 98 | EDN2 | 6719 | 0.342 | -0.1071 | No | ||
| 99 | GIP | 6788 | 0.325 | -0.1102 | No | ||
| 100 | COL4A5 | 6894 | 0.292 | -0.1154 | No | ||
| 101 | SLC6A4 | 6906 | 0.288 | -0.1155 | No | ||
| 102 | BCAT2 | 6944 | 0.280 | -0.1170 | No | ||
| 103 | LYL1 | 7018 | 0.260 | -0.1205 | No | ||
| 104 | RBP2 | 7091 | 0.239 | -0.1240 | No | ||
| 105 | LRP1 | 7097 | 0.238 | -0.1239 | No | ||
| 106 | TRIM47 | 7127 | 0.231 | -0.1250 | No | ||
| 107 | NDFIP2 | 7184 | 0.216 | -0.1277 | No | ||
| 108 | CCK | 7258 | 0.200 | -0.1313 | No | ||
| 109 | CHRNG | 7365 | 0.177 | -0.1368 | No | ||
| 110 | VAV1 | 7385 | 0.169 | -0.1375 | No | ||
| 111 | ST6GAL1 | 7468 | 0.147 | -0.1417 | No | ||
| 112 | NBL1 | 7484 | 0.145 | -0.1423 | No | ||
| 113 | 1200015F23RIK | 7557 | 0.128 | -0.1460 | No | ||
| 114 | AKR1B7 | 7626 | 0.111 | -0.1495 | No | ||
| 115 | PDE9A | 7694 | 0.092 | -0.1530 | No | ||
| 116 | ATP6V0A2 | 7738 | 0.082 | -0.1552 | No | ||
| 117 | NPR3 | 7759 | 0.077 | -0.1562 | No | ||
| 118 | C1QB | 7867 | 0.050 | -0.1619 | No | ||
| 119 | PPARA | 7960 | 0.023 | -0.1669 | No | ||
| 120 | CCNA1 | 8008 | 0.012 | -0.1695 | No | ||
| 121 | CLCN2 | 8027 | 0.008 | -0.1705 | No | ||
| 122 | MAPRE3 | 8086 | -0.005 | -0.1736 | No | ||
| 123 | SLC5A11 | 8134 | -0.015 | -0.1762 | No | ||
| 124 | MTF2 | 8155 | -0.021 | -0.1772 | No | ||
| 125 | GPD1 | 8373 | -0.080 | -0.1889 | No | ||
| 126 | PIPOX | 8410 | -0.089 | -0.1908 | No | ||
| 127 | GRINA | 8491 | -0.112 | -0.1949 | No | ||
| 128 | SST | 8599 | -0.139 | -0.2005 | No | ||
| 129 | RBP4 | 8642 | -0.147 | -0.2026 | No | ||
| 130 | ALDOB | 8667 | -0.153 | -0.2036 | No | ||
| 131 | LSP1 | 8732 | -0.168 | -0.2068 | No | ||
| 132 | IRF4 | 8764 | -0.179 | -0.2081 | No | ||
| 133 | BC052328 | 8831 | -0.195 | -0.2114 | No | ||
| 134 | HIC1 | 8933 | -0.216 | -0.2165 | No | ||
| 135 | MCPT2 | 8937 | -0.217 | -0.2163 | No | ||
| 136 | PCSK5 | 8940 | -0.217 | -0.2160 | No | ||
| 137 | CD22 | 8944 | -0.218 | -0.2158 | No | ||
| 138 | ABCC3 | 8946 | -0.219 | -0.2154 | No | ||
| 139 | LAT2 | 9148 | -0.269 | -0.2259 | No | ||
| 140 | IL4I1 | 9161 | -0.271 | -0.2261 | No | ||
| 141 | 9830147J24RIK | 9184 | -0.276 | -0.2268 | No | ||
| 142 | GALNT10 | 9314 | -0.303 | -0.2333 | No | ||
| 143 | SELPL | 9333 | -0.306 | -0.2337 | No | ||
| 144 | INDO | 9335 | -0.306 | -0.2332 | No | ||
| 145 | NPDC1 | 9381 | -0.315 | -0.2351 | No | ||
| 146 | FABP1 | 9396 | -0.318 | -0.2353 | No | ||
| 147 | CD79A | 9400 | -0.319 | -0.2349 | No | ||
| 148 | SLC5A1 | 9429 | -0.325 | -0.2358 | No | ||
| 149 | THEM2 | 9596 | -0.365 | -0.2442 | No | ||
| 150 | CCL21B | 9742 | -0.399 | -0.2514 | No | ||
| 151 | GHRL | 9800 | -0.415 | -0.2538 | No | ||
| 152 | ICOS | 9832 | -0.423 | -0.2547 | No | ||
| 153 | ITGAX | 9840 | -0.425 | -0.2543 | No | ||
| 154 | MEP1B | 9862 | -0.430 | -0.2547 | No | ||
| 155 | ACOX1 | 9872 | -0.432 | -0.2544 | No | ||
| 156 | MUCDHL | 10043 | -0.473 | -0.2628 | No | ||
| 157 | CD53 | 10110 | -0.490 | -0.2655 | No | ||
| 158 | ST3GAL4 | 10262 | -0.522 | -0.2728 | No | ||
| 159 | ADH4 | 10373 | -0.547 | -0.2778 | No | ||
| 160 | MOGAT2 | 10381 | -0.549 | -0.2772 | No | ||
| 161 | 1810020D17RIK | 10407 | -0.554 | -0.2776 | No | ||
| 162 | DAB1 | 10530 | -0.582 | -0.2832 | No | ||
| 163 | MYH14 | 10552 | -0.586 | -0.2833 | No | ||
| 164 | CXCL13 | 10676 | -0.614 | -0.2889 | No | ||
| 165 | FABP7 | 10714 | -0.622 | -0.2897 | No | ||
| 166 | BMP4 | 10728 | -0.625 | -0.2893 | No | ||
| 167 | CX3CL1 | 10774 | -0.636 | -0.2906 | No | ||
| 168 | IGH-VJ558 | 10782 | -0.637 | -0.2898 | No | ||
| 169 | POLR2I | 10809 | -0.645 | -0.2901 | No | ||
| 170 | ENTPD2 | 10816 | -0.646 | -0.2892 | No | ||
| 171 | TEP1 | 11017 | -0.685 | -0.2989 | No | ||
| 172 | G6PC | 11033 | -0.687 | -0.2985 | No | ||
| 173 | CENPJ | 11179 | -0.721 | -0.3051 | No | ||
| 174 | DDR2 | 11184 | -0.722 | -0.3040 | No | ||
| 175 | DNAJB9 | 11207 | -0.726 | -0.3038 | No | ||
| 176 | GALM | 11212 | -0.727 | -0.3027 | No | ||
| 177 | IFNA1 | 11264 | -0.739 | -0.3042 | No | ||
| 178 | NKIRAS1 | 11306 | -0.746 | -0.3050 | No | ||
| 179 | TGFB1 | 11383 | -0.765 | -0.3078 | No | ||
| 180 | REG3A | 11408 | -0.770 | -0.3077 | No | ||
| 181 | ATP6V0B | 11417 | -0.772 | -0.3067 | No | ||
| 182 | CIDEC | 11424 | -0.773 | -0.3056 | No | ||
| 183 | GABRB1 | 11564 | -0.799 | -0.3118 | No | ||
| 184 | CLCA3 | 11748 | -0.835 | -0.3203 | No | ||
| 185 | CYP27B1 | 11750 | -0.835 | -0.3188 | No | ||
| 186 | BTD | 11755 | -0.836 | -0.3175 | No | ||
| 187 | PPAPDC1A | 11787 | -0.844 | -0.3176 | No | ||
| 188 | SMPD2 | 11807 | -0.850 | -0.3171 | No | ||
| 189 | SLC11A2 | 11965 | -0.880 | -0.3241 | No | ||
| 190 | RDH7 | 11979 | -0.883 | -0.3232 | No | ||
| 191 | EVI2A | 12078 | -0.902 | -0.3269 | No | ||
| 192 | ARSA | 12217 | -0.930 | -0.3327 | No | ||
| 193 | GCG | 12243 | -0.934 | -0.3324 | No | ||
| 194 | TM4SF5 | 12363 | -0.962 | -0.3371 | No | ||
| 195 | REG1 | 12424 | -0.973 | -0.3386 | No | ||
| 196 | MCFD2 | 12432 | -0.974 | -0.3372 | No | ||
| 197 | SLC4A4 | 12532 | -0.996 | -0.3408 | No | ||
| 198 | CXCL14 | 12608 | -1.014 | -0.3431 | No | ||
| 199 | SEPT4 | 12685 | -1.030 | -0.3453 | No | ||
| 200 | TLL1 | 12890 | -1.068 | -0.3545 | No | ||
| 201 | GDI1 | 12922 | -1.077 | -0.3543 | No | ||
| 202 | 1810010M01RIK | 13116 | -1.115 | -0.3628 | No | ||
| 203 | CYP4F13 | 13147 | -1.121 | -0.3624 | No | ||
| 204 | NAPSA | 13337 | -1.166 | -0.3706 | No | ||
| 205 | TAX1BP3 | 13360 | -1.171 | -0.3696 | No | ||
| 206 | ABCC2 | 13557 | -1.219 | -0.3781 | No | ||
| 207 | RHOC | 13629 | -1.238 | -0.3797 | No | ||
| 208 | LTBP4 | 13689 | -1.254 | -0.3807 | No | ||
| 209 | HAGH | 13704 | -1.257 | -0.3791 | No | ||
| 210 | CASP7 | 13756 | -1.268 | -0.3796 | No | ||
| 211 | GPX4 | 13774 | -1.275 | -0.3782 | No | ||
| 212 | CD79B | 13826 | -1.290 | -0.3786 | No | ||
| 213 | ST8SIA4 | 13855 | -1.296 | -0.3778 | No | ||
| 214 | PCOLCE | 13953 | -1.317 | -0.3807 | No | ||
| 215 | STK10 | 13980 | -1.322 | -0.3797 | No | ||
| 216 | NFIA | 14031 | -1.334 | -0.3800 | No | ||
| 217 | CD3E | 14126 | -1.358 | -0.3826 | Yes | ||
| 218 | IL12RB2 | 14140 | -1.362 | -0.3808 | Yes | ||
| 219 | SLC35B1 | 14168 | -1.369 | -0.3798 | Yes | ||
| 220 | DPEP1 | 14169 | -1.369 | -0.3773 | Yes | ||
| 221 | BASP1 | 14179 | -1.373 | -0.3753 | Yes | ||
| 222 | INPP5D | 14237 | -1.387 | -0.3758 | Yes | ||
| 223 | COL18A1 | 14256 | -1.392 | -0.3743 | Yes | ||
| 224 | CDKN2D | 14271 | -1.395 | -0.3725 | Yes | ||
| 225 | OGDH | 14295 | -1.403 | -0.3712 | Yes | ||
| 226 | SLPI | 14345 | -1.417 | -0.3713 | Yes | ||
| 227 | CD72 | 14379 | -1.426 | -0.3705 | Yes | ||
| 228 | TMPRSS8 | 14452 | -1.442 | -0.3717 | Yes | ||
| 229 | UPP1 | 14499 | -1.455 | -0.3716 | Yes | ||
| 230 | A230106M15RIK | 14520 | -1.461 | -0.3700 | Yes | ||
| 231 | PLTP | 14543 | -1.468 | -0.3685 | Yes | ||
| 232 | ALB1 | 14550 | -1.471 | -0.3662 | Yes | ||
| 233 | KCNJ11 | 14617 | -1.488 | -0.3670 | Yes | ||
| 234 | TCF21 | 14627 | -1.491 | -0.3648 | Yes | ||
| 235 | SOD3 | 14691 | -1.506 | -0.3655 | Yes | ||
| 236 | CEACAM10 | 14746 | -1.520 | -0.3657 | Yes | ||
| 237 | IL12RB1 | 14857 | -1.550 | -0.3688 | Yes | ||
| 238 | CYP2S1 | 14933 | -1.573 | -0.3701 | Yes | ||
| 239 | IKBKE | 14955 | -1.579 | -0.3683 | Yes | ||
| 240 | PTPRE | 15023 | -1.599 | -0.3691 | Yes | ||
| 241 | RUNX2 | 15043 | -1.605 | -0.3672 | Yes | ||
| 242 | NR1I3 | 15050 | -1.606 | -0.3645 | Yes | ||
| 243 | IGK-V1 | 15064 | -1.610 | -0.3623 | Yes | ||
| 244 | ABHD8 | 15078 | -1.616 | -0.3600 | Yes | ||
| 245 | GGTLA1 | 15187 | -1.650 | -0.3629 | Yes | ||
| 246 | SLC2A5 | 15227 | -1.663 | -0.3620 | Yes | ||
| 247 | PCX | 15233 | -1.665 | -0.3592 | Yes | ||
| 248 | SOX18 | 15264 | -1.673 | -0.3578 | Yes | ||
| 249 | KLRB1C | 15317 | -1.692 | -0.3576 | Yes | ||
| 250 | SLC27A4 | 15372 | -1.707 | -0.3574 | Yes | ||
| 251 | SIRT2 | 15390 | -1.712 | -0.3552 | Yes | ||
| 252 | IGL-V1 | 15425 | -1.724 | -0.3539 | Yes | ||
| 253 | GSTM1 | 15467 | -1.735 | -0.3529 | Yes | ||
| 254 | SLC2A2 | 15538 | -1.754 | -0.3536 | Yes | ||
| 255 | GNAZ | 15549 | -1.757 | -0.3509 | Yes | ||
| 256 | MCPT1 | 15616 | -1.780 | -0.3512 | Yes | ||
| 257 | ATP6V0A1 | 15730 | -1.829 | -0.3541 | Yes | ||
| 258 | GSTT1 | 15741 | -1.833 | -0.3513 | Yes | ||
| 259 | VDR | 15817 | -1.865 | -0.3519 | Yes | ||
| 260 | CD7 | 15884 | -1.890 | -0.3521 | Yes | ||
| 261 | H2-K1 | 15906 | -1.900 | -0.3498 | Yes | ||
| 262 | RDH5 | 15915 | -1.903 | -0.3467 | Yes | ||
| 263 | NR1H3 | 15951 | -1.919 | -0.3451 | Yes | ||
| 264 | FLT3L | 15952 | -1.919 | -0.3416 | Yes | ||
| 265 | CIDEB | 15975 | -1.928 | -0.3393 | Yes | ||
| 266 | CYP2C40 | 15982 | -1.931 | -0.3360 | Yes | ||
| 267 | CNTNAP1 | 16052 | -1.964 | -0.3362 | Yes | ||
| 268 | KEAP1 | 16069 | -1.970 | -0.3335 | Yes | ||
| 269 | SIPA1 | 16080 | -1.973 | -0.3304 | Yes | ||
| 270 | VAV2 | 16126 | -1.992 | -0.3292 | Yes | ||
| 271 | GZMA | 16132 | -1.995 | -0.3258 | Yes | ||
| 272 | PAFAH2 | 16186 | -2.022 | -0.3250 | Yes | ||
| 273 | CHPT1 | 16217 | -2.031 | -0.3230 | Yes | ||
| 274 | CRAT | 16218 | -2.031 | -0.3192 | Yes | ||
| 275 | SULT2B1 | 16219 | -2.032 | -0.3155 | Yes | ||
| 276 | SOAT2 | 16296 | -2.065 | -0.3159 | Yes | ||
| 277 | CPT2 | 16330 | -2.081 | -0.3139 | Yes | ||
| 278 | DGAT1 | 16420 | -2.124 | -0.3149 | Yes | ||
| 279 | ACY3 | 16470 | -2.145 | -0.3136 | Yes | ||
| 280 | CD3G | 16489 | -2.154 | -0.3106 | Yes | ||
| 281 | ITGAE | 16518 | -2.168 | -0.3082 | Yes | ||
| 282 | PRLR | 16611 | -2.220 | -0.3092 | Yes | ||
| 283 | DHRS1 | 16704 | -2.264 | -0.3101 | Yes | ||
| 284 | GSTA4 | 16884 | -2.371 | -0.3155 | Yes | ||
| 285 | PRRG2 | 16894 | -2.376 | -0.3116 | Yes | ||
| 286 | USP2 | 16921 | -2.392 | -0.3087 | Yes | ||
| 287 | ITGB2 | 16926 | -2.399 | -0.3045 | Yes | ||
| 288 | OGN | 16943 | -2.411 | -0.3010 | Yes | ||
| 289 | SLC35C2 | 16988 | -2.440 | -0.2989 | Yes | ||
| 290 | DPT | 16997 | -2.446 | -0.2949 | Yes | ||
| 291 | AGR2 | 17036 | -2.470 | -0.2924 | Yes | ||
| 292 | IRF1 | 17041 | -2.472 | -0.2881 | Yes | ||
| 293 | ARHGDIB | 17066 | -2.488 | -0.2848 | Yes | ||
| 294 | CTSW | 17093 | -2.508 | -0.2817 | Yes | ||
| 295 | ITGB7 | 17105 | -2.513 | -0.2777 | Yes | ||
| 296 | PIK3CD | 17239 | -2.615 | -0.2802 | Yes | ||
| 297 | MYT1L | 17307 | -2.669 | -0.2789 | Yes | ||
| 298 | CD3D | 17351 | -2.704 | -0.2763 | Yes | ||
| 299 | ACE | 17483 | -2.804 | -0.2784 | Yes | ||
| 300 | PDZK1 | 17513 | -2.828 | -0.2748 | Yes | ||
| 301 | IGH-6 | 17613 | -2.925 | -0.2748 | Yes | ||
| 302 | TCTA | 17662 | -2.985 | -0.2720 | Yes | ||
| 303 | ITGAL | 17671 | -2.992 | -0.2669 | Yes | ||
| 304 | FGD3 | 17709 | -3.034 | -0.2634 | Yes | ||
| 305 | CYB5R3 | 17713 | -3.036 | -0.2580 | Yes | ||
| 306 | SPHK2 | 17761 | -3.087 | -0.2549 | Yes | ||
| 307 | RFXANK | 17762 | -3.087 | -0.2492 | Yes | ||
| 308 | PSMB8 | 17796 | -3.119 | -0.2453 | Yes | ||
| 309 | BC011209 | 17819 | -3.150 | -0.2408 | Yes | ||
| 310 | FMNL1 | 17840 | -3.173 | -0.2360 | Yes | ||
| 311 | RGS10 | 17851 | -3.191 | -0.2307 | Yes | ||
| 312 | ACP5 | 17927 | -3.297 | -0.2288 | Yes | ||
| 313 | S100A13 | 17951 | -3.333 | -0.2240 | Yes | ||
| 314 | MGST2 | 17963 | -3.350 | -0.2184 | Yes | ||
| 315 | GPR89A | 17995 | -3.399 | -0.2139 | Yes | ||
| 316 | GPSM3 | 18020 | -3.450 | -0.2089 | Yes | ||
| 317 | ABCB9 | 18043 | -3.490 | -0.2037 | Yes | ||
| 318 | EPHX2 | 18060 | -3.512 | -0.1981 | Yes | ||
| 319 | WDTC1 | 18119 | -3.634 | -0.1946 | Yes | ||
| 320 | GNA11 | 18147 | -3.679 | -0.1894 | Yes | ||
| 321 | CORO1A | 18156 | -3.694 | -0.1830 | Yes | ||
| 322 | CST7 | 18184 | -3.739 | -0.1777 | Yes | ||
| 323 | ABCD1 | 18235 | -3.874 | -0.1733 | Yes | ||
| 324 | CD2 | 18264 | -3.962 | -0.1676 | Yes | ||
| 325 | TCAP | 18387 | -4.376 | -0.1662 | Yes | ||
| 326 | IL2RB | 18402 | -4.418 | -0.1589 | Yes | ||
| 327 | VAMP5 | 18404 | -4.430 | -0.1508 | Yes | ||
| 328 | GNPDA1 | 18416 | -4.535 | -0.1431 | Yes | ||
| 329 | ZAP70 | 18442 | -4.631 | -0.1360 | Yes | ||
| 330 | PPAP2A | 18454 | -4.676 | -0.1280 | Yes | ||
| 331 | ANXA6 | 18465 | -4.759 | -0.1199 | Yes | ||
| 332 | POU2AF1 | 18469 | -4.782 | -0.1113 | Yes | ||
| 333 | PRPSAP1 | 18471 | -4.787 | -0.1025 | Yes | ||
| 334 | RAC2 | 18475 | -4.810 | -0.0939 | Yes | ||
| 335 | CAPN3 | 18510 | -5.012 | -0.0866 | Yes | ||
| 336 | CD82 | 18525 | -5.141 | -0.0779 | Yes | ||
| 337 | CD160 | 18559 | -5.571 | -0.0695 | Yes | ||
| 338 | CD6 | 18561 | -5.587 | -0.0593 | Yes | ||
| 339 | ATP2A3 | 18562 | -5.589 | -0.0491 | Yes | ||
| 340 | ADAM19 | 18584 | -6.172 | -0.0389 | Yes | ||
| 341 | CD5 | 18602 | -6.983 | -0.0270 | Yes | ||
| 342 | MOV10 | 18603 | -7.025 | -0.0142 | Yes | ||
| 343 | DTX1 | 18614 | -8.089 | 0.0001 | Yes |
