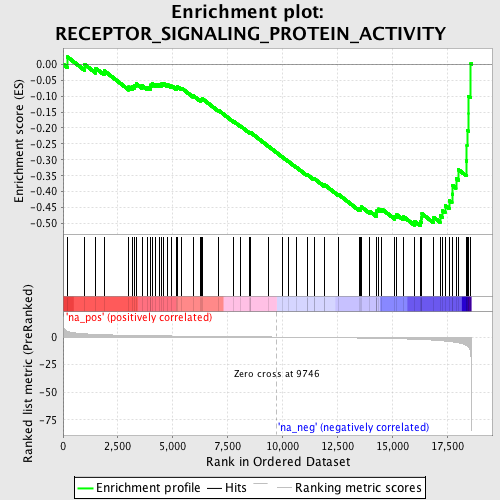
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | RECEPTOR_SIGNALING_PROTEIN_ACTIVITY |
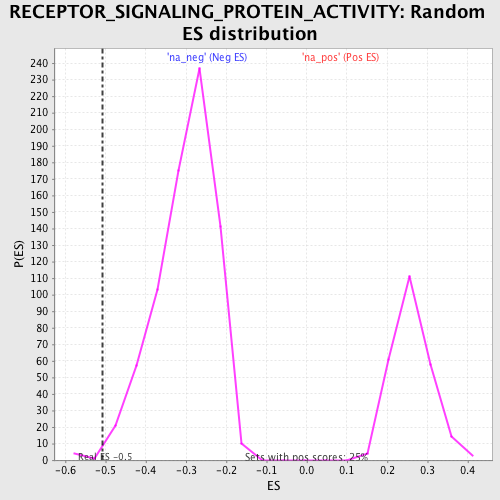
| Enrichment Score (ES) | -0.5075778 |
| Normalized Enrichment Score (NES) | -1.680363 |
| Nominal p-value | 0.0066755675 |
| FDR q-value | 0.6151847 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TBL3 | 193 | 5.258 | 0.0230 | No | ||
| 2 | MAPK10 | 983 | 2.944 | -0.0008 | No | ||
| 3 | TYROBP | 1488 | 2.355 | -0.0130 | No | ||
| 4 | BAG1 | 1871 | 2.081 | -0.0204 | No | ||
| 5 | IFITM1 | 3000 | 1.545 | -0.0714 | No | ||
| 6 | TYRO3 | 3165 | 1.485 | -0.0708 | No | ||
| 7 | MAPK13 | 3249 | 1.457 | -0.0660 | No | ||
| 8 | MAP3K10 | 3326 | 1.427 | -0.0610 | No | ||
| 9 | IL18BP | 3599 | 1.327 | -0.0672 | No | ||
| 10 | IRAK1 | 3833 | 1.257 | -0.0718 | No | ||
| 11 | MAP3K13 | 3979 | 1.212 | -0.0719 | No | ||
| 12 | MAPK8 | 3997 | 1.205 | -0.0652 | No | ||
| 13 | MAP3K6 | 4064 | 1.188 | -0.0612 | No | ||
| 14 | MAP3K7 | 4229 | 1.146 | -0.0627 | No | ||
| 15 | BLNK | 4374 | 1.108 | -0.0635 | No | ||
| 16 | MAPK7 | 4474 | 1.083 | -0.0619 | No | ||
| 17 | ACVR1C | 4554 | 1.064 | -0.0594 | No | ||
| 18 | ERBB2 | 4752 | 1.016 | -0.0636 | No | ||
| 19 | IL1F5 | 4953 | 0.968 | -0.0682 | No | ||
| 20 | CSPG4 | 5182 | 0.919 | -0.0746 | No | ||
| 21 | ACVR1 | 5221 | 0.905 | -0.0709 | No | ||
| 22 | LYN | 5405 | 0.860 | -0.0753 | No | ||
| 23 | AMHR2 | 5921 | 0.754 | -0.0983 | No | ||
| 24 | GNAZ | 6281 | 0.682 | -0.1133 | No | ||
| 25 | PPARGC1B | 6312 | 0.674 | -0.1107 | No | ||
| 26 | MAP3K9 | 6348 | 0.665 | -0.1083 | No | ||
| 27 | CD19 | 7072 | 0.516 | -0.1440 | No | ||
| 28 | MAP3K4 | 7784 | 0.379 | -0.1800 | No | ||
| 29 | SMAD6 | 8080 | 0.321 | -0.1938 | No | ||
| 30 | SKIL | 8501 | 0.244 | -0.2149 | No | ||
| 31 | ACVRL1 | 8523 | 0.238 | -0.2146 | No | ||
| 32 | ARF1 | 8524 | 0.238 | -0.2130 | No | ||
| 33 | ACVR2A | 9382 | 0.081 | -0.2587 | No | ||
| 34 | NCR1 | 9992 | -0.057 | -0.2912 | No | ||
| 35 | IL1RL1 | 10282 | -0.122 | -0.3060 | No | ||
| 36 | IL1RN | 10641 | -0.195 | -0.3241 | No | ||
| 37 | CCL4 | 11116 | -0.294 | -0.3478 | No | ||
| 38 | SMAD7 | 11130 | -0.297 | -0.3466 | No | ||
| 39 | KIT | 11438 | -0.360 | -0.3609 | No | ||
| 40 | SP110 | 11456 | -0.365 | -0.3595 | No | ||
| 41 | ACVR2B | 11893 | -0.462 | -0.3800 | No | ||
| 42 | NSMAF | 11904 | -0.465 | -0.3776 | No | ||
| 43 | MAPK4 | 12548 | -0.616 | -0.4084 | No | ||
| 44 | MAP3K5 | 13494 | -0.867 | -0.4538 | No | ||
| 45 | ACVR1B | 13573 | -0.893 | -0.4524 | No | ||
| 46 | TGFBR1 | 13591 | -0.902 | -0.4475 | No | ||
| 47 | INSR | 13977 | -1.022 | -0.4618 | No | ||
| 48 | MAPK12 | 14274 | -1.127 | -0.4706 | No | ||
| 49 | ALK | 14282 | -1.130 | -0.4638 | No | ||
| 50 | IRAK2 | 14301 | -1.134 | -0.4576 | No | ||
| 51 | BAG4 | 14377 | -1.160 | -0.4542 | No | ||
| 52 | MAPK14 | 14531 | -1.214 | -0.4548 | No | ||
| 53 | IRS1 | 15115 | -1.449 | -0.4770 | No | ||
| 54 | MAPK1 | 15209 | -1.490 | -0.4725 | No | ||
| 55 | PLCE1 | 15502 | -1.645 | -0.4778 | No | ||
| 56 | NLK | 16010 | -1.942 | -0.4928 | No | ||
| 57 | MAP4K1 | 16285 | -2.168 | -0.4938 | Yes | ||
| 58 | SHC1 | 16333 | -2.217 | -0.4822 | Yes | ||
| 59 | ARNT | 16352 | -2.227 | -0.4690 | Yes | ||
| 60 | MAPK9 | 16893 | -2.804 | -0.4803 | Yes | ||
| 61 | MAP3K11 | 17183 | -3.201 | -0.4756 | Yes | ||
| 62 | DAXX | 17296 | -3.389 | -0.4601 | Yes | ||
| 63 | SOCS2 | 17411 | -3.609 | -0.4433 | Yes | ||
| 64 | IKBKE | 17597 | -3.998 | -0.4278 | Yes | ||
| 65 | CDK5 | 17726 | -4.322 | -0.4072 | Yes | ||
| 66 | MAPK11 | 17757 | -4.401 | -0.3809 | Yes | ||
| 67 | STAT3 | 17907 | -4.850 | -0.3581 | Yes | ||
| 68 | MAP3K3 | 18023 | -5.201 | -0.3312 | Yes | ||
| 69 | TGFBR2 | 18385 | -7.438 | -0.3034 | Yes | ||
| 70 | CD3E | 18405 | -7.633 | -0.2558 | Yes | ||
| 71 | STAT2 | 18412 | -7.685 | -0.2073 | Yes | ||
| 72 | CDKN1B | 18474 | -8.692 | -0.1553 | Yes | ||
| 73 | STAT1 | 18476 | -8.708 | -0.1000 | Yes | ||
| 74 | IL4R | 18586 | -16.906 | 0.0016 | Yes |