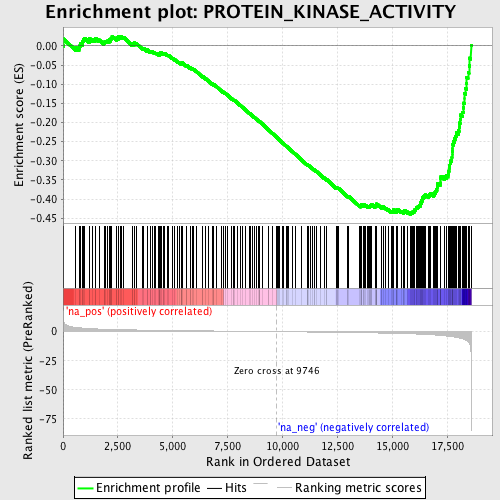
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | PROTEIN_KINASE_ACTIVITY |
| Enrichment Score (ES) | -0.44025275 |
| Normalized Enrichment Score (NES) | -1.6461117 |
| Nominal p-value | 0.0011273957 |
| FDR q-value | 0.6207768 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RIPK4 | 17 | 9.178 | 0.0183 | No | ||
| 2 | CSNK1D | 578 | 3.646 | -0.0046 | No | ||
| 3 | EPHB2 | 726 | 3.367 | -0.0055 | No | ||
| 4 | CDC42BPB | 755 | 3.314 | -0.0001 | No | ||
| 5 | MAP3K7IP1 | 779 | 3.271 | 0.0055 | No | ||
| 6 | RYK | 871 | 3.118 | 0.0070 | No | ||
| 7 | MERTK | 894 | 3.079 | 0.0123 | No | ||
| 8 | MARK1 | 920 | 3.043 | 0.0173 | No | ||
| 9 | MAPK10 | 983 | 2.944 | 0.0201 | No | ||
| 10 | NEK4 | 1189 | 2.677 | 0.0145 | No | ||
| 11 | PRKCZ | 1203 | 2.659 | 0.0194 | No | ||
| 12 | RPS6KA1 | 1354 | 2.492 | 0.0164 | No | ||
| 13 | CSF1R | 1457 | 2.392 | 0.0158 | No | ||
| 14 | PDGFRA | 1473 | 2.373 | 0.0200 | No | ||
| 15 | TWF1 | 1643 | 2.236 | 0.0155 | No | ||
| 16 | RPS6KA3 | 1863 | 2.086 | 0.0079 | No | ||
| 17 | PDGFRB | 1899 | 2.068 | 0.0103 | No | ||
| 18 | FGFR1 | 1948 | 2.038 | 0.0120 | No | ||
| 19 | TTK | 2008 | 2.006 | 0.0130 | No | ||
| 20 | MAPKAPK2 | 2095 | 1.954 | 0.0124 | No | ||
| 21 | CCL2 | 2098 | 1.953 | 0.0164 | No | ||
| 22 | GTF2H1 | 2175 | 1.914 | 0.0162 | No | ||
| 23 | IRAK3 | 2179 | 1.913 | 0.0201 | No | ||
| 24 | CHEK2 | 2226 | 1.886 | 0.0215 | No | ||
| 25 | EFNA4 | 2227 | 1.886 | 0.0254 | No | ||
| 26 | NRP2 | 2425 | 1.787 | 0.0185 | No | ||
| 27 | EIF2AK2 | 2452 | 1.774 | 0.0208 | No | ||
| 28 | STK3 | 2512 | 1.750 | 0.0212 | No | ||
| 29 | FLT4 | 2528 | 1.743 | 0.0240 | No | ||
| 30 | RIPK1 | 2636 | 1.696 | 0.0218 | No | ||
| 31 | EPHA5 | 2654 | 1.689 | 0.0244 | No | ||
| 32 | PTK7 | 2748 | 1.644 | 0.0227 | No | ||
| 33 | TESK1 | 3157 | 1.487 | 0.0036 | No | ||
| 34 | TYRO3 | 3165 | 1.485 | 0.0064 | No | ||
| 35 | EPGN | 3245 | 1.458 | 0.0051 | No | ||
| 36 | MAPK13 | 3249 | 1.457 | 0.0080 | No | ||
| 37 | MAP3K10 | 3326 | 1.427 | 0.0068 | No | ||
| 38 | TTN | 3638 | 1.316 | -0.0073 | No | ||
| 39 | PLK4 | 3662 | 1.309 | -0.0058 | No | ||
| 40 | PDK4 | 3826 | 1.260 | -0.0121 | No | ||
| 41 | IRAK1 | 3833 | 1.257 | -0.0098 | No | ||
| 42 | MAP3K13 | 3979 | 1.212 | -0.0151 | No | ||
| 43 | MAPK8 | 3997 | 1.205 | -0.0135 | No | ||
| 44 | MAP3K6 | 4064 | 1.188 | -0.0147 | No | ||
| 45 | ROR1 | 4171 | 1.160 | -0.0180 | No | ||
| 46 | MAP3K7 | 4229 | 1.146 | -0.0187 | No | ||
| 47 | NEK11 | 4330 | 1.119 | -0.0218 | No | ||
| 48 | CSNK1A1 | 4389 | 1.104 | -0.0227 | No | ||
| 49 | DAPK2 | 4400 | 1.100 | -0.0209 | No | ||
| 50 | RPS6KC1 | 4409 | 1.098 | -0.0190 | No | ||
| 51 | PDGFRL | 4425 | 1.095 | -0.0176 | No | ||
| 52 | MAPK7 | 4474 | 1.083 | -0.0179 | No | ||
| 53 | ACVR1C | 4554 | 1.064 | -0.0200 | No | ||
| 54 | ROCK2 | 4595 | 1.057 | -0.0199 | No | ||
| 55 | MAP2K7 | 4635 | 1.048 | -0.0199 | No | ||
| 56 | ERBB2 | 4752 | 1.016 | -0.0241 | No | ||
| 57 | ROR2 | 4799 | 1.005 | -0.0245 | No | ||
| 58 | AXL | 4976 | 0.961 | -0.0320 | No | ||
| 59 | DDR1 | 5075 | 0.941 | -0.0354 | No | ||
| 60 | ACVR1 | 5221 | 0.905 | -0.0414 | No | ||
| 61 | STK19 | 5319 | 0.880 | -0.0448 | No | ||
| 62 | CSNK2B | 5403 | 0.861 | -0.0476 | No | ||
| 63 | LYN | 5405 | 0.860 | -0.0458 | No | ||
| 64 | NEK2 | 5414 | 0.859 | -0.0445 | No | ||
| 65 | DDR2 | 5444 | 0.852 | -0.0442 | No | ||
| 66 | STK38L | 5605 | 0.820 | -0.0512 | No | ||
| 67 | CDK10 | 5617 | 0.817 | -0.0501 | No | ||
| 68 | TLK1 | 5802 | 0.779 | -0.0585 | No | ||
| 69 | VRK1 | 5806 | 0.779 | -0.0571 | No | ||
| 70 | PRKAG3 | 5888 | 0.761 | -0.0599 | No | ||
| 71 | TSSK2 | 5945 | 0.750 | -0.0614 | No | ||
| 72 | CDK9 | 6057 | 0.727 | -0.0659 | No | ||
| 73 | MAP3K9 | 6348 | 0.665 | -0.0803 | No | ||
| 74 | HUNK | 6350 | 0.664 | -0.0789 | No | ||
| 75 | PRKCE | 6499 | 0.633 | -0.0857 | No | ||
| 76 | DAPK3 | 6616 | 0.612 | -0.0907 | No | ||
| 77 | PRKAG1 | 6793 | 0.571 | -0.0991 | No | ||
| 78 | CHUK | 6859 | 0.560 | -0.1015 | No | ||
| 79 | AKT1 | 6874 | 0.557 | -0.1011 | No | ||
| 80 | TIE1 | 6982 | 0.535 | -0.1058 | No | ||
| 81 | CDK7 | 7199 | 0.492 | -0.1165 | No | ||
| 82 | TAOK2 | 7288 | 0.474 | -0.1203 | No | ||
| 83 | PAK2 | 7318 | 0.467 | -0.1209 | No | ||
| 84 | EXOSC10 | 7379 | 0.456 | -0.1232 | No | ||
| 85 | ERBB4 | 7485 | 0.436 | -0.1280 | No | ||
| 86 | CDC42BPA | 7689 | 0.399 | -0.1382 | No | ||
| 87 | LTK | 7783 | 0.379 | -0.1425 | No | ||
| 88 | MAP3K4 | 7784 | 0.379 | -0.1417 | No | ||
| 89 | TSSK3 | 7824 | 0.371 | -0.1431 | No | ||
| 90 | NTRK1 | 7931 | 0.348 | -0.1481 | No | ||
| 91 | MAPK6 | 8076 | 0.322 | -0.1553 | No | ||
| 92 | EIF2AK1 | 8189 | 0.302 | -0.1607 | No | ||
| 93 | BCKDK | 8301 | 0.283 | -0.1662 | No | ||
| 94 | RPS6KB2 | 8511 | 0.241 | -0.1771 | No | ||
| 95 | ACVRL1 | 8523 | 0.238 | -0.1772 | No | ||
| 96 | KDR | 8623 | 0.216 | -0.1821 | No | ||
| 97 | SRPK2 | 8729 | 0.199 | -0.1874 | No | ||
| 98 | TEK | 8819 | 0.185 | -0.1919 | No | ||
| 99 | ERN2 | 8890 | 0.174 | -0.1953 | No | ||
| 100 | TXK | 8933 | 0.166 | -0.1972 | No | ||
| 101 | MAPKAPK3 | 8935 | 0.166 | -0.1970 | No | ||
| 102 | CIT | 8970 | 0.159 | -0.1985 | No | ||
| 103 | EFNA3 | 9076 | 0.138 | -0.2039 | No | ||
| 104 | MAP2K1 | 9344 | 0.087 | -0.2183 | No | ||
| 105 | ACVR2A | 9382 | 0.081 | -0.2201 | No | ||
| 106 | NEK6 | 9542 | 0.046 | -0.2287 | No | ||
| 107 | MET | 9727 | 0.003 | -0.2387 | No | ||
| 108 | MAPKAPK5 | 9746 | -0.000 | -0.2397 | No | ||
| 109 | BRSK2 | 9790 | -0.011 | -0.2420 | No | ||
| 110 | FLT3 | 9819 | -0.016 | -0.2435 | No | ||
| 111 | TRIB1 | 9842 | -0.020 | -0.2446 | No | ||
| 112 | TSSK6 | 10013 | -0.062 | -0.2537 | No | ||
| 113 | MUSK | 10041 | -0.067 | -0.2551 | No | ||
| 114 | RPS6KA5 | 10181 | -0.097 | -0.2624 | No | ||
| 115 | PTK6 | 10202 | -0.102 | -0.2633 | No | ||
| 116 | RIPK3 | 10247 | -0.111 | -0.2655 | No | ||
| 117 | EIF2AK4 | 10279 | -0.121 | -0.2669 | No | ||
| 118 | TSSK1A | 10464 | -0.160 | -0.2766 | No | ||
| 119 | TESK2 | 10584 | -0.183 | -0.2827 | No | ||
| 120 | PRKACB | 10855 | -0.239 | -0.2969 | No | ||
| 121 | CCL4 | 11116 | -0.294 | -0.3104 | No | ||
| 122 | FGFR4 | 11138 | -0.299 | -0.3109 | No | ||
| 123 | WNK4 | 11154 | -0.302 | -0.3111 | No | ||
| 124 | SGK2 | 11190 | -0.309 | -0.3124 | No | ||
| 125 | STK38 | 11295 | -0.330 | -0.3174 | No | ||
| 126 | PAK1 | 11346 | -0.340 | -0.3194 | No | ||
| 127 | KIT | 11438 | -0.360 | -0.3236 | No | ||
| 128 | MKNK2 | 11471 | -0.366 | -0.3246 | No | ||
| 129 | MYLK | 11553 | -0.385 | -0.3282 | No | ||
| 130 | PLK1 | 11717 | -0.422 | -0.3361 | No | ||
| 131 | ACVR2B | 11893 | -0.462 | -0.3447 | No | ||
| 132 | PDK1 | 11983 | -0.481 | -0.3485 | No | ||
| 133 | FRK | 12026 | -0.490 | -0.3498 | No | ||
| 134 | LIMK1 | 12451 | -0.593 | -0.3717 | No | ||
| 135 | WNK3 | 12462 | -0.595 | -0.3710 | No | ||
| 136 | FLT1 | 12483 | -0.600 | -0.3708 | No | ||
| 137 | ERN1 | 12523 | -0.609 | -0.3716 | No | ||
| 138 | MAPK4 | 12548 | -0.616 | -0.3717 | No | ||
| 139 | PHKG2 | 12974 | -0.721 | -0.3933 | No | ||
| 140 | BCR | 12982 | -0.724 | -0.3921 | No | ||
| 141 | CSNK2A2 | 13009 | -0.730 | -0.3920 | No | ||
| 142 | MAP3K5 | 13494 | -0.867 | -0.4166 | No | ||
| 143 | ACVR1B | 13573 | -0.893 | -0.4190 | No | ||
| 144 | TGFBR1 | 13591 | -0.902 | -0.4180 | No | ||
| 145 | RPS6KA2 | 13596 | -0.903 | -0.4163 | No | ||
| 146 | TYK2 | 13602 | -0.905 | -0.4147 | No | ||
| 147 | PRKCB1 | 13692 | -0.927 | -0.4176 | No | ||
| 148 | PHKA1 | 13712 | -0.936 | -0.4167 | No | ||
| 149 | ROCK1 | 13717 | -0.937 | -0.4149 | No | ||
| 150 | MAST1 | 13800 | -0.962 | -0.4174 | No | ||
| 151 | ERBB3 | 13885 | -0.989 | -0.4199 | No | ||
| 152 | TGFA | 13925 | -1.001 | -0.4199 | No | ||
| 153 | EFNB3 | 13968 | -1.019 | -0.4201 | No | ||
| 154 | INSR | 13977 | -1.022 | -0.4184 | No | ||
| 155 | EGFR | 13992 | -1.027 | -0.4170 | No | ||
| 156 | STK11 | 14028 | -1.040 | -0.4167 | No | ||
| 157 | STK16 | 14040 | -1.045 | -0.4151 | No | ||
| 158 | CSNK2A1 | 14044 | -1.046 | -0.4131 | No | ||
| 159 | PRKD1 | 14218 | -1.106 | -0.4202 | No | ||
| 160 | MAP3K2 | 14220 | -1.108 | -0.4179 | No | ||
| 161 | MST1R | 14262 | -1.123 | -0.4178 | No | ||
| 162 | MAPK12 | 14274 | -1.127 | -0.4161 | No | ||
| 163 | ALK | 14282 | -1.130 | -0.4141 | No | ||
| 164 | IRAK2 | 14301 | -1.134 | -0.4127 | No | ||
| 165 | HIPK3 | 14507 | -1.208 | -0.4213 | No | ||
| 166 | MAPK14 | 14531 | -1.214 | -0.4200 | No | ||
| 167 | MAP2K3 | 14582 | -1.229 | -0.4202 | No | ||
| 168 | PDK3 | 14691 | -1.273 | -0.4234 | No | ||
| 169 | CAMKK2 | 14810 | -1.320 | -0.4271 | No | ||
| 170 | COL4A3BP | 14975 | -1.389 | -0.4331 | No | ||
| 171 | PRKCI | 15034 | -1.413 | -0.4333 | No | ||
| 172 | UHMK1 | 15035 | -1.414 | -0.4303 | No | ||
| 173 | MKNK1 | 15044 | -1.418 | -0.4278 | No | ||
| 174 | PBK | 15193 | -1.482 | -0.4328 | No | ||
| 175 | MAPK1 | 15209 | -1.490 | -0.4305 | No | ||
| 176 | VRK2 | 15212 | -1.492 | -0.4274 | No | ||
| 177 | TRIM27 | 15253 | -1.514 | -0.4265 | No | ||
| 178 | MAP4K4 | 15416 | -1.601 | -0.4319 | No | ||
| 179 | CDK2 | 15515 | -1.650 | -0.4338 | No | ||
| 180 | CRIM1 | 15531 | -1.657 | -0.4312 | No | ||
| 181 | CDK3 | 15570 | -1.680 | -0.4297 | No | ||
| 182 | MAP4K5 | 15706 | -1.757 | -0.4334 | No | ||
| 183 | CLK1 | 15833 | -1.834 | -0.4364 | Yes | ||
| 184 | STK4 | 15864 | -1.857 | -0.4342 | Yes | ||
| 185 | PLK3 | 15928 | -1.897 | -0.4336 | Yes | ||
| 186 | CDKL1 | 15959 | -1.912 | -0.4313 | Yes | ||
| 187 | NLK | 16010 | -1.942 | -0.4299 | Yes | ||
| 188 | LMTK2 | 16033 | -1.960 | -0.4270 | Yes | ||
| 189 | AKT2 | 16083 | -1.993 | -0.4255 | Yes | ||
| 190 | GSK3B | 16087 | -1.994 | -0.4215 | Yes | ||
| 191 | IGF1R | 16151 | -2.060 | -0.4206 | Yes | ||
| 192 | LATS2 | 16196 | -2.095 | -0.4186 | Yes | ||
| 193 | ATM | 16233 | -2.123 | -0.4162 | Yes | ||
| 194 | MARK4 | 16276 | -2.162 | -0.4139 | Yes | ||
| 195 | MAP4K1 | 16285 | -2.168 | -0.4098 | Yes | ||
| 196 | TRIB2 | 16336 | -2.218 | -0.4079 | Yes | ||
| 197 | WNK1 | 16341 | -2.221 | -0.4035 | Yes | ||
| 198 | PRKX | 16376 | -2.248 | -0.4006 | Yes | ||
| 199 | PXK | 16388 | -2.261 | -0.3965 | Yes | ||
| 200 | EPHB6 | 16423 | -2.290 | -0.3935 | Yes | ||
| 201 | STK17B | 16471 | -2.334 | -0.3912 | Yes | ||
| 202 | ILK | 16520 | -2.381 | -0.3889 | Yes | ||
| 203 | PRKD3 | 16666 | -2.529 | -0.3915 | Yes | ||
| 204 | SRPK1 | 16686 | -2.540 | -0.3872 | Yes | ||
| 205 | PRKCH | 16734 | -2.592 | -0.3843 | Yes | ||
| 206 | MAPK9 | 16893 | -2.804 | -0.3871 | Yes | ||
| 207 | IGF2R | 16943 | -2.855 | -0.3837 | Yes | ||
| 208 | MAP4K3 | 16972 | -2.892 | -0.3792 | Yes | ||
| 209 | CPNE3 | 17012 | -2.943 | -0.3752 | Yes | ||
| 210 | MINK1 | 17042 | -2.989 | -0.3705 | Yes | ||
| 211 | CSNK1G2 | 17052 | -2.996 | -0.3647 | Yes | ||
| 212 | LIMK2 | 17076 | -3.030 | -0.3596 | Yes | ||
| 213 | MAP3K11 | 17183 | -3.201 | -0.3587 | Yes | ||
| 214 | MAP3K8 | 17204 | -3.241 | -0.3530 | Yes | ||
| 215 | RIPK2 | 17205 | -3.242 | -0.3462 | Yes | ||
| 216 | TLK2 | 17220 | -3.268 | -0.3402 | Yes | ||
| 217 | PHKA2 | 17387 | -3.561 | -0.3418 | Yes | ||
| 218 | CLK2 | 17470 | -3.735 | -0.3384 | Yes | ||
| 219 | MAP2K6 | 17544 | -3.918 | -0.3342 | Yes | ||
| 220 | TNK2 | 17559 | -3.937 | -0.3267 | Yes | ||
| 221 | ULK1 | 17590 | -3.987 | -0.3200 | Yes | ||
| 222 | IKBKE | 17597 | -3.998 | -0.3120 | Yes | ||
| 223 | PINK1 | 17633 | -4.084 | -0.3053 | Yes | ||
| 224 | MAP3K12 | 17675 | -4.196 | -0.2988 | Yes | ||
| 225 | EIF2AK3 | 17721 | -4.310 | -0.2922 | Yes | ||
| 226 | CDK5 | 17726 | -4.322 | -0.2834 | Yes | ||
| 227 | SNF1LK | 17729 | -4.328 | -0.2744 | Yes | ||
| 228 | MAPK11 | 17757 | -4.401 | -0.2667 | Yes | ||
| 229 | PIM1 | 17761 | -4.415 | -0.2576 | Yes | ||
| 230 | PIM2 | 17790 | -4.508 | -0.2497 | Yes | ||
| 231 | ALS2CR2 | 17838 | -4.629 | -0.2426 | Yes | ||
| 232 | CRKL | 17893 | -4.821 | -0.2355 | Yes | ||
| 233 | STK10 | 17918 | -4.882 | -0.2265 | Yes | ||
| 234 | MAP3K3 | 18023 | -5.201 | -0.2213 | Yes | ||
| 235 | CSNK1E | 18048 | -5.309 | -0.2115 | Yes | ||
| 236 | PDPK1 | 18064 | -5.362 | -0.2011 | Yes | ||
| 237 | BRD2 | 18095 | -5.480 | -0.1913 | Yes | ||
| 238 | PDK2 | 18108 | -5.558 | -0.1803 | Yes | ||
| 239 | SNF1LK2 | 18208 | -6.071 | -0.1730 | Yes | ||
| 240 | BRDT | 18244 | -6.288 | -0.1618 | Yes | ||
| 241 | ITK | 18258 | -6.356 | -0.1492 | Yes | ||
| 242 | DYRK1A | 18283 | -6.537 | -0.1368 | Yes | ||
| 243 | MARK2 | 18285 | -6.557 | -0.1231 | Yes | ||
| 244 | SNRK | 18346 | -7.103 | -0.1115 | Yes | ||
| 245 | TGFBR2 | 18385 | -7.438 | -0.0980 | Yes | ||
| 246 | SGK3 | 18397 | -7.586 | -0.0828 | Yes | ||
| 247 | MAP4K2 | 18455 | -8.325 | -0.0685 | Yes | ||
| 248 | CDK5R1 | 18501 | -9.126 | -0.0518 | Yes | ||
| 249 | PTK2B | 18534 | -10.458 | -0.0317 | Yes | ||
| 250 | TEC | 18590 | -17.252 | 0.0014 | Yes |
