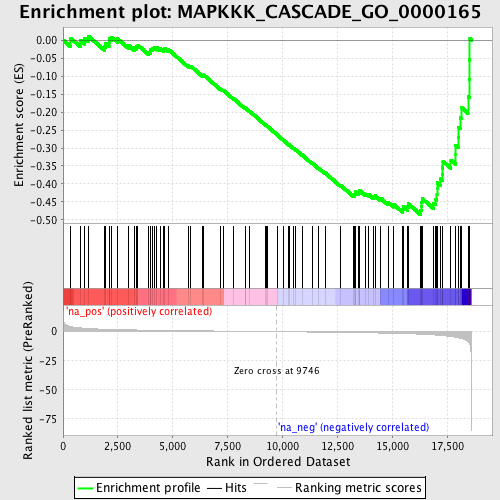
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MAPKKK_CASCADE_GO_0000165 |
| Enrichment Score (ES) | -0.48482344 |
| Normalized Enrichment Score (NES) | -1.6432656 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.59945166 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SOD1 | 342 | 4.350 | 0.0055 | No | ||
| 2 | MAP3K7IP1 | 779 | 3.271 | -0.0001 | No | ||
| 3 | MAPK10 | 983 | 2.944 | 0.0051 | No | ||
| 4 | GPS1 | 1151 | 2.730 | 0.0111 | No | ||
| 5 | NRTN | 1881 | 2.075 | -0.0168 | No | ||
| 6 | FGFR1 | 1948 | 2.038 | -0.0091 | No | ||
| 7 | MAPKAPK2 | 2095 | 1.954 | -0.0063 | No | ||
| 8 | MBIP | 2102 | 1.950 | 0.0041 | No | ||
| 9 | GADD45G | 2212 | 1.894 | 0.0087 | No | ||
| 10 | MAP2K4 | 2476 | 1.763 | 0.0041 | No | ||
| 11 | FGFR3 | 2991 | 1.548 | -0.0151 | No | ||
| 12 | EPGN | 3245 | 1.458 | -0.0207 | No | ||
| 13 | MAP3K10 | 3326 | 1.427 | -0.0172 | No | ||
| 14 | EDA2R | 3411 | 1.394 | -0.0141 | No | ||
| 15 | EGF | 3908 | 1.234 | -0.0340 | No | ||
| 16 | MAP3K13 | 3979 | 1.212 | -0.0312 | No | ||
| 17 | MAPK8 | 3997 | 1.205 | -0.0254 | No | ||
| 18 | MAP3K6 | 4064 | 1.188 | -0.0225 | No | ||
| 19 | ATP6AP2 | 4151 | 1.165 | -0.0207 | No | ||
| 20 | TDGF1 | 4277 | 1.137 | -0.0212 | No | ||
| 21 | CD24 | 4417 | 1.096 | -0.0227 | No | ||
| 22 | DUSP9 | 4565 | 1.062 | -0.0248 | No | ||
| 23 | MAP2K7 | 4635 | 1.048 | -0.0227 | No | ||
| 24 | SCG2 | 4811 | 1.003 | -0.0267 | No | ||
| 25 | CHRNA7 | 5708 | 0.798 | -0.0706 | No | ||
| 26 | ADORA2B | 5827 | 0.775 | -0.0728 | No | ||
| 27 | MAP3K9 | 6348 | 0.665 | -0.0972 | No | ||
| 28 | C5AR1 | 6409 | 0.652 | -0.0968 | No | ||
| 29 | TPD52L1 | 7174 | 0.496 | -0.1353 | No | ||
| 30 | TAOK2 | 7288 | 0.474 | -0.1388 | No | ||
| 31 | MAP3K4 | 7784 | 0.379 | -0.1635 | No | ||
| 32 | DUSP4 | 8297 | 0.283 | -0.1895 | No | ||
| 33 | GNG3 | 8304 | 0.282 | -0.1883 | No | ||
| 34 | ADRA2A | 8499 | 0.244 | -0.1974 | No | ||
| 35 | ADRA2C | 9208 | 0.115 | -0.2350 | No | ||
| 36 | TRAF7 | 9273 | 0.102 | -0.2379 | No | ||
| 37 | PIK3CB | 9318 | 0.093 | -0.2398 | No | ||
| 38 | RGS4 | 9781 | -0.009 | -0.2647 | No | ||
| 39 | GRM4 | 10046 | -0.068 | -0.2785 | No | ||
| 40 | HIPK2 | 10294 | -0.124 | -0.2912 | No | ||
| 41 | SPRED1 | 10327 | -0.132 | -0.2922 | No | ||
| 42 | DBNL | 10498 | -0.168 | -0.3004 | No | ||
| 43 | FPR1 | 10598 | -0.186 | -0.3048 | No | ||
| 44 | MAPK8IP2 | 10890 | -0.247 | -0.3191 | No | ||
| 45 | PAK1 | 11346 | -0.340 | -0.3418 | No | ||
| 46 | FGF2 | 11646 | -0.406 | -0.3557 | No | ||
| 47 | PROK2 | 11938 | -0.471 | -0.3688 | No | ||
| 48 | MDFI | 12642 | -0.637 | -0.4033 | No | ||
| 49 | DUSP8 | 13227 | -0.791 | -0.4304 | No | ||
| 50 | DUSP22 | 13301 | -0.811 | -0.4299 | No | ||
| 51 | DUSP10 | 13310 | -0.814 | -0.4259 | No | ||
| 52 | EDG5 | 13319 | -0.815 | -0.4218 | No | ||
| 53 | CARTPT | 13458 | -0.855 | -0.4246 | No | ||
| 54 | MAP3K5 | 13494 | -0.867 | -0.4217 | No | ||
| 55 | ADRB3 | 13525 | -0.879 | -0.4185 | No | ||
| 56 | GADD45B | 13797 | -0.961 | -0.4278 | No | ||
| 57 | TGFA | 13925 | -1.001 | -0.4292 | No | ||
| 58 | NF1 | 14157 | -1.082 | -0.4357 | No | ||
| 59 | MAP3K2 | 14220 | -1.108 | -0.4329 | No | ||
| 60 | GHRL | 14477 | -1.198 | -0.4402 | No | ||
| 61 | CAMKK2 | 14810 | -1.320 | -0.4508 | No | ||
| 62 | SH2D3C | 15058 | -1.425 | -0.4563 | No | ||
| 63 | CXCR4 | 15483 | -1.636 | -0.4702 | No | ||
| 64 | PLCE1 | 15502 | -1.645 | -0.4622 | No | ||
| 65 | MAP4K5 | 15706 | -1.757 | -0.4634 | No | ||
| 66 | CD81 | 15734 | -1.776 | -0.4551 | No | ||
| 67 | MAP4K1 | 16285 | -2.168 | -0.4729 | Yes | ||
| 68 | GPS2 | 16325 | -2.206 | -0.4629 | Yes | ||
| 69 | SHC1 | 16333 | -2.217 | -0.4511 | Yes | ||
| 70 | PKN1 | 16361 | -2.238 | -0.4402 | Yes | ||
| 71 | MAPK9 | 16893 | -2.804 | -0.4535 | Yes | ||
| 72 | MAP4K3 | 16972 | -2.892 | -0.4418 | Yes | ||
| 73 | DUSP2 | 17039 | -2.987 | -0.4289 | Yes | ||
| 74 | MINK1 | 17042 | -2.989 | -0.4126 | Yes | ||
| 75 | MAPK8IP3 | 17046 | -2.994 | -0.3963 | Yes | ||
| 76 | MAP3K11 | 17183 | -3.201 | -0.3860 | Yes | ||
| 77 | ADRB2 | 17284 | -3.362 | -0.3729 | Yes | ||
| 78 | DAXX | 17296 | -3.389 | -0.3549 | Yes | ||
| 79 | RGS3 | 17313 | -3.419 | -0.3370 | Yes | ||
| 80 | MAP3K12 | 17675 | -4.196 | -0.3334 | Yes | ||
| 81 | CRKL | 17893 | -4.821 | -0.3186 | Yes | ||
| 82 | FGF13 | 17897 | -4.828 | -0.2922 | Yes | ||
| 83 | CCM2 | 18012 | -5.158 | -0.2700 | Yes | ||
| 84 | MAP3K3 | 18023 | -5.201 | -0.2419 | Yes | ||
| 85 | SPRED2 | 18094 | -5.476 | -0.2156 | Yes | ||
| 86 | CD27 | 18151 | -5.749 | -0.1870 | Yes | ||
| 87 | MAP4K2 | 18455 | -8.325 | -0.1576 | Yes | ||
| 88 | DUSP16 | 18515 | -9.556 | -0.1082 | Yes | ||
| 89 | MADD | 18522 | -9.863 | -0.0543 | Yes | ||
| 90 | DUSP6 | 18542 | -10.788 | 0.0040 | Yes |