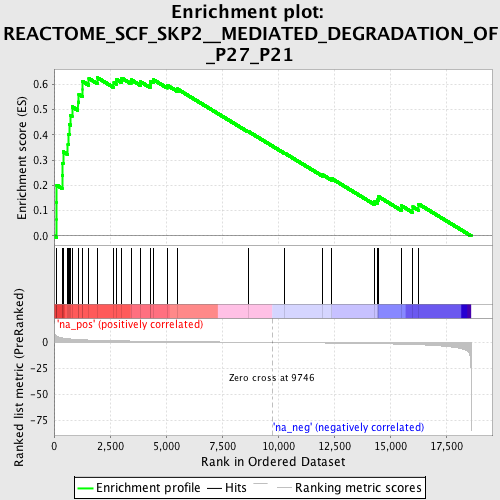
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_SCF_SKP2__MEDIATED_DEGRADATION_OF_P27_P21 |
| Enrichment Score (ES) | 0.6260548 |
| Normalized Enrichment Score (NES) | 2.1733086 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.4637566E-4 |
| FWER p-Value | 0.0020 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMD3 | 113 | 6.052 | 0.0643 | Yes | ||
| 2 | PSMC5 | 116 | 5.927 | 0.1331 | Yes | ||
| 3 | PSMB1 | 122 | 5.824 | 0.2005 | Yes | ||
| 4 | PSMD14 | 359 | 4.294 | 0.2378 | Yes | ||
| 5 | PSMD8 | 375 | 4.251 | 0.2864 | Yes | ||
| 6 | PSMB2 | 403 | 4.126 | 0.3329 | Yes | ||
| 7 | PSME3 | 617 | 3.558 | 0.3628 | Yes | ||
| 8 | PSMC3 | 636 | 3.532 | 0.4029 | Yes | ||
| 9 | PSMA7 | 691 | 3.421 | 0.4398 | Yes | ||
| 10 | PSMD12 | 727 | 3.364 | 0.4770 | Yes | ||
| 11 | PSMA4 | 803 | 3.234 | 0.5106 | Yes | ||
| 12 | CDKN1A | 1066 | 2.845 | 0.5296 | Yes | ||
| 13 | PSMC6 | 1106 | 2.791 | 0.5599 | Yes | ||
| 14 | PSMB3 | 1273 | 2.563 | 0.5808 | Yes | ||
| 15 | CUL1 | 1275 | 2.562 | 0.6106 | Yes | ||
| 16 | PSMB6 | 1554 | 2.298 | 0.6223 | Yes | ||
| 17 | PSMA3 | 1928 | 2.048 | 0.6261 | Yes | ||
| 18 | PSMC1 | 2671 | 1.682 | 0.6057 | No | ||
| 19 | PSMC2 | 2773 | 1.635 | 0.6193 | No | ||
| 20 | PSMA5 | 3019 | 1.536 | 0.6239 | No | ||
| 21 | PSMA6 | 3436 | 1.384 | 0.6176 | No | ||
| 22 | PSMB10 | 3857 | 1.251 | 0.6096 | No | ||
| 23 | PSMD7 | 4289 | 1.132 | 0.5995 | No | ||
| 24 | PSMA2 | 4304 | 1.128 | 0.6119 | No | ||
| 25 | PSMB5 | 4427 | 1.094 | 0.6181 | No | ||
| 26 | PSMD5 | 5045 | 0.949 | 0.5959 | No | ||
| 27 | PSMA1 | 5497 | 0.840 | 0.5814 | No | ||
| 28 | PSMD2 | 8658 | 0.211 | 0.4138 | No | ||
| 29 | PSMB9 | 10303 | -0.125 | 0.3267 | No | ||
| 30 | PSMC4 | 11992 | -0.483 | 0.2415 | No | ||
| 31 | PSMD11 | 12400 | -0.579 | 0.2263 | No | ||
| 32 | PSMB4 | 14300 | -1.134 | 0.1373 | No | ||
| 33 | PSMD9 | 14452 | -1.187 | 0.1430 | No | ||
| 34 | PSME1 | 14492 | -1.203 | 0.1549 | No | ||
| 35 | CDK2 | 15515 | -1.650 | 0.1190 | No | ||
| 36 | PSMD10 | 16018 | -1.944 | 0.1146 | No | ||
| 37 | PSMD4 | 16275 | -2.162 | 0.1260 | No |