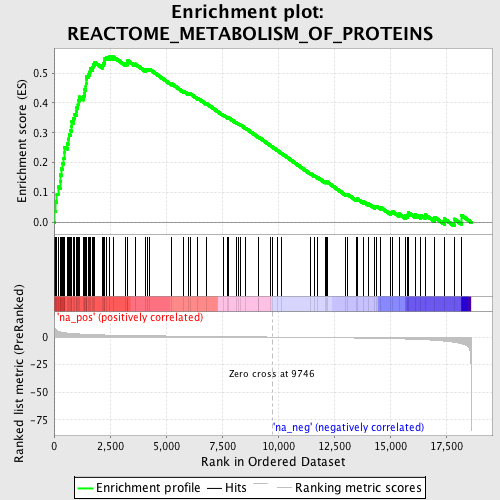
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_METABOLISM_OF_PROTEINS |
| Enrichment Score (ES) | 0.5545911 |
| Normalized Enrichment Score (NES) | 2.2568944 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EIF2B5 | 38 | 7.912 | 0.0372 | Yes | ||
| 2 | ACTB | 81 | 6.729 | 0.0683 | Yes | ||
| 3 | EIF4EBP1 | 126 | 5.803 | 0.0947 | Yes | ||
| 4 | EEF1D | 177 | 5.373 | 0.1186 | Yes | ||
| 5 | RPS3 | 270 | 4.716 | 0.1370 | Yes | ||
| 6 | EIF4B | 291 | 4.621 | 0.1588 | Yes | ||
| 7 | F2 | 323 | 4.441 | 0.1792 | Yes | ||
| 8 | CCT6A | 372 | 4.253 | 0.1977 | Yes | ||
| 9 | PABPC1 | 419 | 4.086 | 0.2154 | Yes | ||
| 10 | RPS19 | 455 | 3.961 | 0.2332 | Yes | ||
| 11 | RPS3A | 462 | 3.936 | 0.2524 | Yes | ||
| 12 | RPL4 | 590 | 3.612 | 0.2634 | Yes | ||
| 13 | EIF2S2 | 623 | 3.545 | 0.2793 | Yes | ||
| 14 | DPM1 | 664 | 3.462 | 0.2943 | Yes | ||
| 15 | RPL30 | 722 | 3.372 | 0.3079 | Yes | ||
| 16 | EIF4A1 | 769 | 3.290 | 0.3217 | Yes | ||
| 17 | CCT4 | 792 | 3.257 | 0.3367 | Yes | ||
| 18 | EIF4E | 869 | 3.123 | 0.3481 | Yes | ||
| 19 | RPL27A | 905 | 3.065 | 0.3614 | Yes | ||
| 20 | CCT3 | 984 | 2.944 | 0.3718 | Yes | ||
| 21 | CCT8 | 1014 | 2.909 | 0.3846 | Yes | ||
| 22 | ETF1 | 1065 | 2.849 | 0.3960 | Yes | ||
| 23 | EEF1G | 1072 | 2.838 | 0.4098 | Yes | ||
| 24 | PFDN6 | 1133 | 2.756 | 0.4202 | Yes | ||
| 25 | F10 | 1325 | 2.520 | 0.4224 | Yes | ||
| 26 | RPS24 | 1352 | 2.493 | 0.4333 | Yes | ||
| 27 | EEF2 | 1372 | 2.472 | 0.4446 | Yes | ||
| 28 | EIF2B1 | 1420 | 2.431 | 0.4541 | Yes | ||
| 29 | GPAA1 | 1423 | 2.429 | 0.4660 | Yes | ||
| 30 | EIF2B2 | 1429 | 2.424 | 0.4778 | Yes | ||
| 31 | RPS2 | 1435 | 2.416 | 0.4895 | Yes | ||
| 32 | RPL14 | 1526 | 2.321 | 0.4961 | Yes | ||
| 33 | VBP1 | 1593 | 2.272 | 0.5038 | Yes | ||
| 34 | GAS6 | 1604 | 2.266 | 0.5145 | Yes | ||
| 35 | RPS23 | 1706 | 2.194 | 0.5199 | Yes | ||
| 36 | PROC | 1740 | 2.168 | 0.5289 | Yes | ||
| 37 | RPL38 | 1798 | 2.120 | 0.5363 | Yes | ||
| 38 | EIF2S1 | 2158 | 1.922 | 0.5265 | Yes | ||
| 39 | PLAUR | 2199 | 1.904 | 0.5338 | Yes | ||
| 40 | RPSA | 2258 | 1.867 | 0.5399 | Yes | ||
| 41 | CCT2 | 2266 | 1.862 | 0.5487 | Yes | ||
| 42 | TBCA | 2345 | 1.829 | 0.5536 | Yes | ||
| 43 | DHPS | 2489 | 1.759 | 0.5546 | Yes | ||
| 44 | CCT5 | 2648 | 1.691 | 0.5544 | No | ||
| 45 | F7 | 3174 | 1.483 | 0.5334 | No | ||
| 46 | RPS4X | 3291 | 1.442 | 0.5343 | No | ||
| 47 | EIF4G1 | 3294 | 1.441 | 0.5414 | No | ||
| 48 | RPS15A | 3613 | 1.323 | 0.5308 | No | ||
| 49 | RPLP2 | 4087 | 1.182 | 0.5111 | No | ||
| 50 | GSPT2 | 4159 | 1.164 | 0.5130 | No | ||
| 51 | RPL5 | 4255 | 1.142 | 0.5135 | No | ||
| 52 | EIF5 | 5246 | 0.898 | 0.4645 | No | ||
| 53 | RPLP1 | 5791 | 0.781 | 0.4390 | No | ||
| 54 | RPL18A | 5980 | 0.740 | 0.4325 | No | ||
| 55 | TBCE | 6086 | 0.721 | 0.4305 | No | ||
| 56 | RPS21 | 6418 | 0.651 | 0.4158 | No | ||
| 57 | RPL8 | 6789 | 0.572 | 0.3987 | No | ||
| 58 | RPL27 | 7570 | 0.420 | 0.3586 | No | ||
| 59 | SEMA6D | 7747 | 0.387 | 0.3510 | No | ||
| 60 | RPS29 | 7801 | 0.376 | 0.3500 | No | ||
| 61 | PFDN1 | 8136 | 0.310 | 0.3335 | No | ||
| 62 | RPS17 | 8241 | 0.293 | 0.3294 | No | ||
| 63 | F9 | 8331 | 0.276 | 0.3260 | No | ||
| 64 | RPS13 | 8557 | 0.231 | 0.3149 | No | ||
| 65 | RPS25 | 9122 | 0.131 | 0.2851 | No | ||
| 66 | PIGN | 9124 | 0.130 | 0.2857 | No | ||
| 67 | RPL6 | 9665 | 0.017 | 0.2567 | No | ||
| 68 | RPL39 | 9744 | 0.000 | 0.2524 | No | ||
| 69 | DPM2 | 9949 | -0.046 | 0.2417 | No | ||
| 70 | PROZ | 10168 | -0.095 | 0.2304 | No | ||
| 71 | RPL35A | 11457 | -0.365 | 0.1626 | No | ||
| 72 | PIGW | 11620 | -0.400 | 0.1558 | No | ||
| 73 | RPL28 | 11770 | -0.432 | 0.1499 | No | ||
| 74 | RPL17 | 12110 | -0.508 | 0.1342 | No | ||
| 75 | RPL41 | 12137 | -0.513 | 0.1353 | No | ||
| 76 | RPS28 | 12184 | -0.526 | 0.1354 | No | ||
| 77 | FURIN | 13017 | -0.732 | 0.0941 | No | ||
| 78 | RPS15 | 13107 | -0.763 | 0.0931 | No | ||
| 79 | PIGF | 13496 | -0.868 | 0.0764 | No | ||
| 80 | RPS14 | 13536 | -0.882 | 0.0787 | No | ||
| 81 | RPS16 | 13812 | -0.967 | 0.0687 | No | ||
| 82 | RPL18 | 14034 | -1.043 | 0.0619 | No | ||
| 83 | EIF5B | 14318 | -1.141 | 0.0523 | No | ||
| 84 | RPS11 | 14399 | -1.166 | 0.0537 | No | ||
| 85 | RPL37A | 14574 | -1.227 | 0.0504 | No | ||
| 86 | EIF4H | 15031 | -1.412 | 0.0328 | No | ||
| 87 | RPL11 | 15108 | -1.448 | 0.0359 | No | ||
| 88 | PIGP | 15397 | -1.592 | 0.0282 | No | ||
| 89 | FAU | 15681 | -1.737 | 0.0215 | No | ||
| 90 | PROS1 | 15794 | -1.813 | 0.0245 | No | ||
| 91 | RPS20 | 15807 | -1.822 | 0.0329 | No | ||
| 92 | RPL19 | 16118 | -2.024 | 0.0262 | No | ||
| 93 | RPS5 | 16374 | -2.246 | 0.0235 | No | ||
| 94 | RPL37 | 16560 | -2.420 | 0.0255 | No | ||
| 95 | PIGA | 16973 | -2.892 | 0.0176 | No | ||
| 96 | PIGH | 17420 | -3.621 | 0.0115 | No | ||
| 97 | TBCC | 17862 | -4.723 | 0.0111 | No | ||
| 98 | GGCX | 18187 | -5.965 | 0.0232 | No |