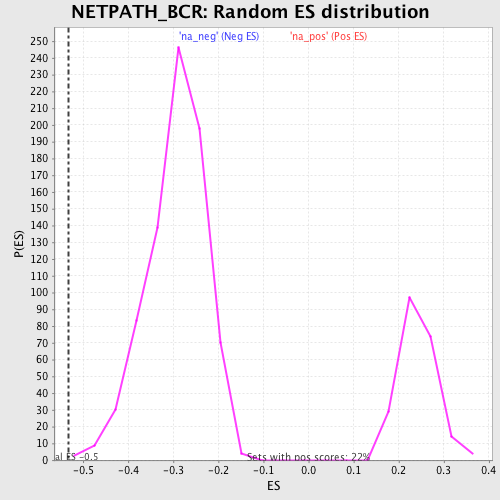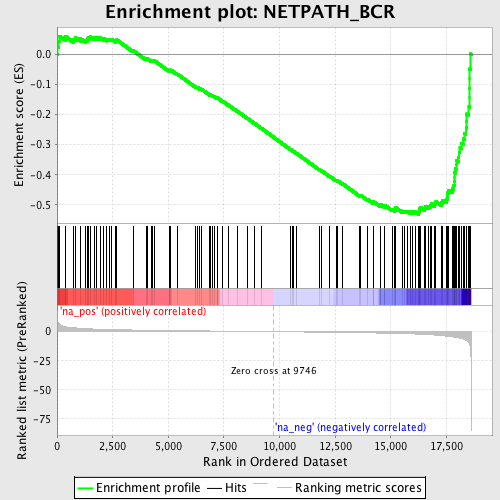
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_absentNotch_versus_normalThy |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | -0.5317454 |
| Normalized Enrichment Score (NES) | -1.8117706 |
| Nominal p-value | 0.0025575447 |
| FDR q-value | 0.12985262 |
| FWER p-Value | 0.858 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BAX | 28 | 8.257 | 0.0239 | No | ||
| 2 | PTK2 | 64 | 7.089 | 0.0438 | No | ||
| 3 | CDK4 | 111 | 6.134 | 0.0602 | No | ||
| 4 | FCGR2B | 362 | 4.281 | 0.0599 | No | ||
| 5 | GRB2 | 736 | 3.346 | 0.0500 | No | ||
| 6 | CRK | 816 | 3.207 | 0.0556 | No | ||
| 7 | RPS6 | 1042 | 2.877 | 0.0523 | No | ||
| 8 | ITPR2 | 1283 | 2.557 | 0.0472 | No | ||
| 9 | RPS6KA1 | 1354 | 2.492 | 0.0511 | No | ||
| 10 | PTPRC | 1422 | 2.430 | 0.0549 | No | ||
| 11 | PIK3R2 | 1505 | 2.342 | 0.0577 | No | ||
| 12 | PPP3R1 | 1664 | 2.223 | 0.0560 | No | ||
| 13 | CYCS | 1777 | 2.140 | 0.0565 | No | ||
| 14 | BTK | 1933 | 2.046 | 0.0544 | No | ||
| 15 | MAPKAPK2 | 2095 | 1.954 | 0.0517 | No | ||
| 16 | NCK1 | 2241 | 1.878 | 0.0497 | No | ||
| 17 | CCNE1 | 2346 | 1.829 | 0.0497 | No | ||
| 18 | LAT2 | 2432 | 1.784 | 0.0506 | No | ||
| 19 | PLCG1 | 2622 | 1.699 | 0.0456 | No | ||
| 20 | CD22 | 2682 | 1.675 | 0.0476 | No | ||
| 21 | PRKCD | 3412 | 1.394 | 0.0125 | No | ||
| 22 | MAPK8 | 3997 | 1.205 | -0.0154 | No | ||
| 23 | DOK3 | 4053 | 1.190 | -0.0147 | No | ||
| 24 | MAP3K7 | 4229 | 1.146 | -0.0206 | No | ||
| 25 | RPS6KB1 | 4288 | 1.133 | -0.0203 | No | ||
| 26 | BLNK | 4374 | 1.108 | -0.0215 | No | ||
| 27 | BCL2L11 | 5035 | 0.951 | -0.0542 | No | ||
| 28 | CREB1 | 5058 | 0.945 | -0.0525 | No | ||
| 29 | GAB2 | 5106 | 0.933 | -0.0522 | No | ||
| 30 | LYN | 5405 | 0.860 | -0.0656 | No | ||
| 31 | PIK3R1 | 6233 | 0.693 | -0.1082 | No | ||
| 32 | CD72 | 6306 | 0.676 | -0.1100 | No | ||
| 33 | RASGRP3 | 6416 | 0.651 | -0.1139 | No | ||
| 34 | PRKCE | 6499 | 0.633 | -0.1164 | No | ||
| 35 | CHUK | 6859 | 0.560 | -0.1341 | No | ||
| 36 | AKT1 | 6874 | 0.557 | -0.1331 | No | ||
| 37 | CCNA2 | 6998 | 0.530 | -0.1381 | No | ||
| 38 | CD19 | 7072 | 0.516 | -0.1405 | No | ||
| 39 | CDK6 | 7188 | 0.494 | -0.1452 | No | ||
| 40 | CDK7 | 7199 | 0.492 | -0.1442 | No | ||
| 41 | GSK3A | 7442 | 0.445 | -0.1559 | No | ||
| 42 | BCAR1 | 7688 | 0.399 | -0.1679 | No | ||
| 43 | CD79A | 8111 | 0.315 | -0.1898 | No | ||
| 44 | HCK | 8538 | 0.234 | -0.2121 | No | ||
| 45 | DOK1 | 8858 | 0.179 | -0.2288 | No | ||
| 46 | PIK3AP1 | 9201 | 0.116 | -0.2469 | No | ||
| 47 | JUN | 10501 | -0.169 | -0.3166 | No | ||
| 48 | SOS1 | 10560 | -0.179 | -0.3192 | No | ||
| 49 | ACTR3 | 10622 | -0.191 | -0.3219 | No | ||
| 50 | BLK | 10746 | -0.215 | -0.3279 | No | ||
| 51 | REL | 11773 | -0.433 | -0.3820 | No | ||
| 52 | HCLS1 | 11875 | -0.458 | -0.3860 | No | ||
| 53 | SOS2 | 12257 | -0.546 | -0.4049 | No | ||
| 54 | MAPK4 | 12548 | -0.616 | -0.4187 | No | ||
| 55 | NFATC2 | 12580 | -0.623 | -0.4185 | No | ||
| 56 | CBLB | 12833 | -0.686 | -0.4300 | No | ||
| 57 | SH3BP2 | 13603 | -0.905 | -0.4688 | No | ||
| 58 | PPP3CA | 13649 | -0.916 | -0.4684 | No | ||
| 59 | MAPK3 | 13948 | -1.009 | -0.4814 | No | ||
| 60 | SYK | 14216 | -1.106 | -0.4924 | No | ||
| 61 | PRKD1 | 14218 | -1.106 | -0.4891 | No | ||
| 62 | MAPK14 | 14531 | -1.214 | -0.5022 | No | ||
| 63 | CR2 | 14543 | -1.217 | -0.4990 | No | ||
| 64 | RASA1 | 14704 | -1.279 | -0.5038 | No | ||
| 65 | IKBKB | 14715 | -1.283 | -0.5003 | No | ||
| 66 | RAPGEF1 | 15065 | -1.427 | -0.5148 | No | ||
| 67 | PTPN11 | 15185 | -1.479 | -0.5167 | No | ||
| 68 | PTPN18 | 15205 | -1.486 | -0.5131 | No | ||
| 69 | MAPK1 | 15209 | -1.490 | -0.5087 | No | ||
| 70 | CDK2 | 15515 | -1.650 | -0.5201 | No | ||
| 71 | ZAP70 | 15615 | -1.707 | -0.5202 | No | ||
| 72 | CD81 | 15734 | -1.776 | -0.5211 | No | ||
| 73 | ACTR2 | 15880 | -1.869 | -0.5232 | No | ||
| 74 | PTPN6 | 15968 | -1.915 | -0.5220 | No | ||
| 75 | GSK3B | 16087 | -1.994 | -0.5223 | No | ||
| 76 | RB1 | 16263 | -2.150 | -0.5251 | Yes | ||
| 77 | CASP9 | 16279 | -2.163 | -0.5193 | Yes | ||
| 78 | MAP4K1 | 16285 | -2.168 | -0.5129 | Yes | ||
| 79 | SHC1 | 16333 | -2.217 | -0.5086 | Yes | ||
| 80 | DAPP1 | 16522 | -2.382 | -0.5114 | Yes | ||
| 81 | CSK | 16538 | -2.400 | -0.5049 | Yes | ||
| 82 | NFKBIA | 16685 | -2.540 | -0.5049 | Yes | ||
| 83 | HDAC5 | 16779 | -2.655 | -0.5018 | Yes | ||
| 84 | CCND3 | 16825 | -2.709 | -0.4959 | Yes | ||
| 85 | ATF2 | 16984 | -2.909 | -0.4955 | Yes | ||
| 86 | GTF2I | 17026 | -2.960 | -0.4886 | Yes | ||
| 87 | LCK | 17279 | -3.355 | -0.4919 | Yes | ||
| 88 | VAV2 | 17336 | -3.460 | -0.4843 | Yes | ||
| 89 | RELA | 17490 | -3.802 | -0.4808 | Yes | ||
| 90 | BCL10 | 17534 | -3.900 | -0.4712 | Yes | ||
| 91 | CCND2 | 17556 | -3.932 | -0.4602 | Yes | ||
| 92 | CBL | 17607 | -4.019 | -0.4505 | Yes | ||
| 93 | WAS | 17765 | -4.434 | -0.4454 | Yes | ||
| 94 | LCP2 | 17831 | -4.615 | -0.4347 | Yes | ||
| 95 | PIP5K1C | 17856 | -4.690 | -0.4216 | Yes | ||
| 96 | PRKCQ | 17869 | -4.739 | -0.4076 | Yes | ||
| 97 | NFATC3 | 17871 | -4.741 | -0.3931 | Yes | ||
| 98 | CRKL | 17893 | -4.821 | -0.3794 | Yes | ||
| 99 | IKBKG | 17938 | -4.921 | -0.3666 | Yes | ||
| 100 | VAV1 | 17943 | -4.944 | -0.3516 | Yes | ||
| 101 | FYN | 18056 | -5.334 | -0.3413 | Yes | ||
| 102 | PDPK1 | 18064 | -5.362 | -0.3252 | Yes | ||
| 103 | PDK2 | 18108 | -5.558 | -0.3104 | Yes | ||
| 104 | CARD11 | 18184 | -5.916 | -0.2962 | Yes | ||
| 105 | ITK | 18258 | -6.356 | -0.2806 | Yes | ||
| 106 | CD79B | 18302 | -6.710 | -0.2623 | Yes | ||
| 107 | NEDD9 | 18379 | -7.388 | -0.2437 | Yes | ||
| 108 | NFATC1 | 18404 | -7.630 | -0.2215 | Yes | ||
| 109 | CASP7 | 18410 | -7.664 | -0.1982 | Yes | ||
| 110 | BCL6 | 18511 | -9.435 | -0.1746 | Yes | ||
| 111 | CD5 | 18527 | -10.056 | -0.1444 | Yes | ||
| 112 | GAB1 | 18529 | -10.215 | -0.1131 | Yes | ||
| 113 | PTK2B | 18534 | -10.458 | -0.0811 | Yes | ||
| 114 | BCL2 | 18536 | -10.511 | -0.0488 | Yes | ||
| 115 | TEC | 18590 | -17.252 | 0.0014 | Yes |