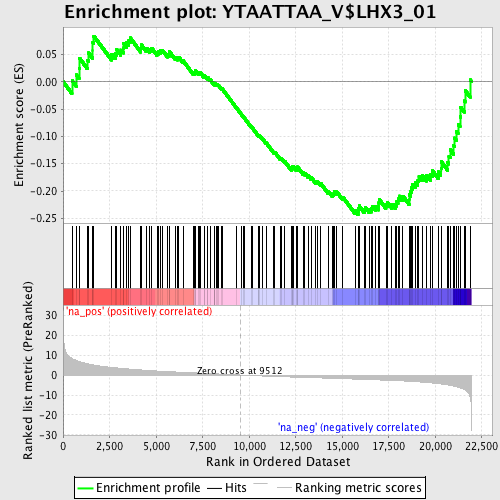
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | YTAATTAA_V$LHX3_01 |
| Enrichment Score (ES) | -0.24299441 |
| Normalized Enrichment Score (NES) | -1.1454741 |
| Nominal p-value | 0.16991644 |
| FDR q-value | 0.5585126 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ARID3B | 479 | 8.275 | 0.0019 | No | ||
| 2 | MTSS1 | 702 | 7.247 | 0.0127 | No | ||
| 3 | ADD3 | 860 | 6.696 | 0.0248 | No | ||
| 4 | H2AFZ | 874 | 6.647 | 0.0434 | No | ||
| 5 | ARNTL | 1310 | 5.592 | 0.0396 | No | ||
| 6 | MLLT10 | 1364 | 5.457 | 0.0530 | No | ||
| 7 | TLE4 | 1595 | 5.061 | 0.0570 | No | ||
| 8 | FLOT1 | 1597 | 5.059 | 0.0716 | No | ||
| 9 | NOTCH2 | 1650 | 4.966 | 0.0835 | No | ||
| 10 | MBNL1 | 2609 | 3.802 | 0.0506 | No | ||
| 11 | XYLT2 | 2801 | 3.608 | 0.0523 | No | ||
| 12 | SRPK2 | 2883 | 3.531 | 0.0587 | No | ||
| 13 | PURA | 3107 | 3.339 | 0.0582 | No | ||
| 14 | CLK2 | 3231 | 3.239 | 0.0619 | No | ||
| 15 | SSBP3 | 3241 | 3.230 | 0.0708 | No | ||
| 16 | STX7 | 3420 | 3.090 | 0.0716 | No | ||
| 17 | ELF4 | 3534 | 2.991 | 0.0750 | No | ||
| 18 | MYST4 | 3603 | 2.947 | 0.0804 | No | ||
| 19 | TYR | 4175 | 2.531 | 0.0615 | No | ||
| 20 | PITPNC1 | 4201 | 2.506 | 0.0676 | No | ||
| 21 | MARCKS | 4488 | 2.329 | 0.0612 | No | ||
| 22 | OPRM1 | 4659 | 2.223 | 0.0599 | No | ||
| 23 | KCNK2 | 4761 | 2.164 | 0.0615 | No | ||
| 24 | CDYL | 5046 | 2.007 | 0.0542 | No | ||
| 25 | DIXDC1 | 5128 | 1.963 | 0.0562 | No | ||
| 26 | RORB | 5234 | 1.911 | 0.0569 | No | ||
| 27 | SEMA5B | 5326 | 1.864 | 0.0581 | No | ||
| 28 | DSPP | 5611 | 1.721 | 0.0501 | No | ||
| 29 | PPM1E | 5694 | 1.688 | 0.0512 | No | ||
| 30 | CYLD | 5710 | 1.680 | 0.0553 | No | ||
| 31 | GNAO1 | 6037 | 1.526 | 0.0448 | No | ||
| 32 | OSBPL8 | 6150 | 1.474 | 0.0439 | No | ||
| 33 | TAL1 | 6215 | 1.442 | 0.0452 | No | ||
| 34 | BHLHB3 | 6445 | 1.339 | 0.0385 | No | ||
| 35 | WNT7A | 7008 | 1.078 | 0.0159 | No | ||
| 36 | MS4A1 | 7055 | 1.058 | 0.0168 | No | ||
| 37 | LRRC4 | 7086 | 1.044 | 0.0184 | No | ||
| 38 | NLGN2 | 7107 | 1.035 | 0.0205 | No | ||
| 39 | NFYA | 7281 | 0.960 | 0.0154 | No | ||
| 40 | HRK | 7307 | 0.948 | 0.0170 | No | ||
| 41 | BNC2 | 7370 | 0.923 | 0.0168 | No | ||
| 42 | RAX | 7580 | 0.836 | 0.0096 | No | ||
| 43 | PRDM1 | 7584 | 0.833 | 0.0119 | No | ||
| 44 | LRRTM1 | 7742 | 0.763 | 0.0069 | No | ||
| 45 | ITPR3 | 7763 | 0.754 | 0.0081 | No | ||
| 46 | PDZRN4 | 7902 | 0.702 | 0.0038 | No | ||
| 47 | USH3A | 8122 | 0.610 | -0.0044 | No | ||
| 48 | PRKAB1 | 8148 | 0.596 | -0.0039 | No | ||
| 49 | IRX4 | 8152 | 0.595 | -0.0023 | No | ||
| 50 | LMO3 | 8247 | 0.558 | -0.0050 | No | ||
| 51 | SSPN | 8292 | 0.542 | -0.0054 | No | ||
| 52 | FZD10 | 8345 | 0.519 | -0.0063 | No | ||
| 53 | MYF6 | 8525 | 0.443 | -0.0132 | No | ||
| 54 | NRL | 8537 | 0.439 | -0.0125 | No | ||
| 55 | CHST8 | 9313 | 0.100 | -0.0477 | No | ||
| 56 | IRX5 | 9571 | -0.029 | -0.0594 | No | ||
| 57 | SP8 | 9690 | -0.081 | -0.0646 | No | ||
| 58 | VAMP8 | 9766 | -0.115 | -0.0677 | No | ||
| 59 | CALN1 | 10102 | -0.264 | -0.0823 | No | ||
| 60 | PPP1R1B | 10155 | -0.280 | -0.0839 | No | ||
| 61 | IRX6 | 10512 | -0.408 | -0.0990 | No | ||
| 62 | LEMD1 | 10519 | -0.412 | -0.0981 | No | ||
| 63 | CGA | 10539 | -0.419 | -0.0978 | No | ||
| 64 | TBR1 | 10724 | -0.484 | -0.1048 | No | ||
| 65 | SALL3 | 10912 | -0.553 | -0.1118 | No | ||
| 66 | HOXC4 | 11324 | -0.689 | -0.1287 | No | ||
| 67 | NKX6-1 | 11378 | -0.709 | -0.1290 | No | ||
| 68 | DLC1 | 11686 | -0.820 | -0.1407 | No | ||
| 69 | LMX1A | 11726 | -0.835 | -0.1401 | No | ||
| 70 | FEV | 11900 | -0.889 | -0.1455 | No | ||
| 71 | DHX38 | 12291 | -1.004 | -0.1605 | No | ||
| 72 | TBX19 | 12303 | -1.007 | -0.1581 | No | ||
| 73 | KLF14 | 12304 | -1.008 | -0.1552 | No | ||
| 74 | LDB2 | 12355 | -1.023 | -0.1545 | No | ||
| 75 | FIGN | 12549 | -1.080 | -0.1602 | No | ||
| 76 | ADAM11 | 12585 | -1.091 | -0.1587 | No | ||
| 77 | PRDM8 | 12593 | -1.092 | -0.1558 | No | ||
| 78 | DMRTA1 | 12902 | -1.187 | -0.1665 | No | ||
| 79 | TRPM8 | 12996 | -1.217 | -0.1673 | No | ||
| 80 | MID1 | 13180 | -1.269 | -0.1720 | No | ||
| 81 | WNT8B | 13322 | -1.308 | -0.1747 | No | ||
| 82 | RTN4RL1 | 13572 | -1.386 | -0.1821 | No | ||
| 83 | EN1 | 13659 | -1.411 | -0.1820 | No | ||
| 84 | SLC6A5 | 13826 | -1.458 | -0.1854 | No | ||
| 85 | ITGA8 | 14252 | -1.581 | -0.2003 | No | ||
| 86 | LRFN5 | 14484 | -1.641 | -0.2062 | No | ||
| 87 | DLL4 | 14548 | -1.664 | -0.2042 | No | ||
| 88 | SULF1 | 14569 | -1.671 | -0.2003 | No | ||
| 89 | KCTD15 | 14702 | -1.713 | -0.2014 | No | ||
| 90 | SLC24A2 | 15026 | -1.808 | -0.2110 | No | ||
| 91 | SH3BGRL2 | 15696 | -2.031 | -0.2358 | No | ||
| 92 | LRRC15 | 15853 | -2.080 | -0.2370 | Yes | ||
| 93 | COL12A1 | 15878 | -2.087 | -0.2321 | Yes | ||
| 94 | CD36 | 15901 | -2.095 | -0.2270 | Yes | ||
| 95 | RGS8 | 16179 | -2.174 | -0.2334 | Yes | ||
| 96 | PROK2 | 16232 | -2.196 | -0.2295 | Yes | ||
| 97 | FIP1L1 | 16468 | -2.279 | -0.2337 | Yes | ||
| 98 | RPA2 | 16574 | -2.316 | -0.2318 | Yes | ||
| 99 | CDH10 | 16631 | -2.333 | -0.2276 | Yes | ||
| 100 | DMD | 16796 | -2.392 | -0.2283 | Yes | ||
| 101 | MAB21L1 | 16941 | -2.445 | -0.2278 | Yes | ||
| 102 | SULF2 | 16966 | -2.454 | -0.2218 | Yes | ||
| 103 | TECTA | 16974 | -2.457 | -0.2150 | Yes | ||
| 104 | SHOX2 | 17350 | -2.597 | -0.2247 | Yes | ||
| 105 | NPR3 | 17430 | -2.622 | -0.2208 | Yes | ||
| 106 | HHIP | 17668 | -2.713 | -0.2238 | Yes | ||
| 107 | HESX1 | 17852 | -2.792 | -0.2241 | Yes | ||
| 108 | RORA | 17932 | -2.824 | -0.2196 | Yes | ||
| 109 | OSR2 | 17997 | -2.853 | -0.2143 | Yes | ||
| 110 | PACSIN3 | 18067 | -2.885 | -0.2091 | Yes | ||
| 111 | PRRX1 | 18256 | -2.957 | -0.2092 | Yes | ||
| 112 | MYT1 | 18594 | -3.122 | -0.2156 | Yes | ||
| 113 | ZFHX1B | 18599 | -3.124 | -0.2068 | Yes | ||
| 114 | BAZ1A | 18669 | -3.160 | -0.2008 | Yes | ||
| 115 | PAX3 | 18724 | -3.198 | -0.1941 | Yes | ||
| 116 | SEMA3A | 18783 | -3.226 | -0.1874 | Yes | ||
| 117 | BACE2 | 18931 | -3.314 | -0.1846 | Yes | ||
| 118 | NDST4 | 19063 | -3.389 | -0.1808 | Yes | ||
| 119 | ELMO1 | 19121 | -3.431 | -0.1735 | Yes | ||
| 120 | IER3 | 19295 | -3.536 | -0.1712 | Yes | ||
| 121 | PIK3R3 | 19528 | -3.708 | -0.1712 | Yes | ||
| 122 | LBX1 | 19742 | -3.891 | -0.1697 | Yes | ||
| 123 | INPPL1 | 19844 | -3.988 | -0.1628 | Yes | ||
| 124 | FZD7 | 20160 | -4.299 | -0.1649 | Yes | ||
| 125 | SEZ6L | 20304 | -4.450 | -0.1586 | Yes | ||
| 126 | NEO1 | 20318 | -4.470 | -0.1462 | Yes | ||
| 127 | BLMH | 20668 | -4.955 | -0.1479 | Yes | ||
| 128 | ELP4 | 20731 | -5.081 | -0.1361 | Yes | ||
| 129 | LMO4 | 20804 | -5.199 | -0.1244 | Yes | ||
| 130 | RREB1 | 20991 | -5.466 | -0.1171 | Yes | ||
| 131 | TFDP2 | 21031 | -5.552 | -0.1029 | Yes | ||
| 132 | SFRS7 | 21128 | -5.770 | -0.0906 | Yes | ||
| 133 | GNA13 | 21224 | -6.051 | -0.0775 | Yes | ||
| 134 | ADAMTS10 | 21329 | -6.371 | -0.0639 | Yes | ||
| 135 | JDP2 | 21351 | -6.434 | -0.0463 | Yes | ||
| 136 | SUCLG2 | 21556 | -7.140 | -0.0350 | Yes | ||
| 137 | PECR | 21603 | -7.342 | -0.0159 | Yes | ||
| 138 | PRKAR2A | 21871 | -10.955 | 0.0035 | Yes |
