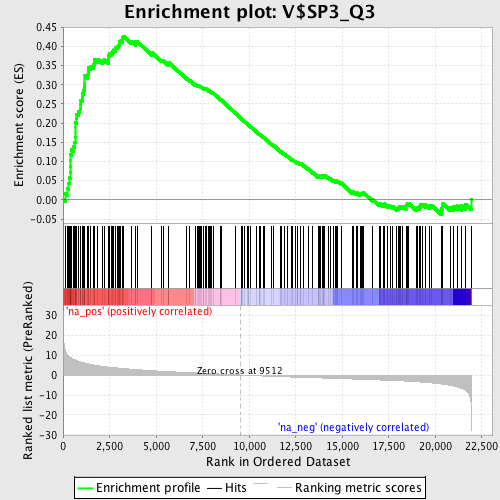
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$SP3_Q3 |
| Enrichment Score (ES) | 0.42679498 |
| Normalized Enrichment Score (NES) | 1.9170092 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009396264 |
| FWER p-Value | 0.041 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ZBTB9 | 118 | 12.444 | 0.0166 | Yes | ||
| 2 | MAML1 | 218 | 10.355 | 0.0304 | Yes | ||
| 3 | CRY1 | 301 | 9.519 | 0.0434 | Yes | ||
| 4 | TBCC | 339 | 9.185 | 0.0580 | Yes | ||
| 5 | JUNB | 376 | 8.916 | 0.0721 | Yes | ||
| 6 | CHES1 | 390 | 8.863 | 0.0872 | Yes | ||
| 7 | YWHAE | 396 | 8.835 | 0.1026 | Yes | ||
| 8 | POFUT1 | 399 | 8.796 | 0.1180 | Yes | ||
| 9 | RNF40 | 439 | 8.481 | 0.1313 | Yes | ||
| 10 | POLR2A | 578 | 7.767 | 0.1387 | Yes | ||
| 11 | PLEKHM1 | 617 | 7.626 | 0.1504 | Yes | ||
| 12 | LDHB | 657 | 7.455 | 0.1618 | Yes | ||
| 13 | SLC4A2 | 665 | 7.427 | 0.1746 | Yes | ||
| 14 | CTDSP1 | 666 | 7.427 | 0.1878 | Yes | ||
| 15 | RASGRP2 | 668 | 7.421 | 0.2008 | Yes | ||
| 16 | PRDM10 | 706 | 7.234 | 0.2119 | Yes | ||
| 17 | CDAN1 | 732 | 7.127 | 0.2234 | Yes | ||
| 18 | RAB11B | 821 | 6.811 | 0.2314 | Yes | ||
| 19 | DET1 | 907 | 6.568 | 0.2391 | Yes | ||
| 20 | FBS1 | 937 | 6.504 | 0.2493 | Yes | ||
| 21 | DNAJC7 | 951 | 6.459 | 0.2601 | Yes | ||
| 22 | FOXP1 | 1045 | 6.223 | 0.2669 | Yes | ||
| 23 | ZBTB4 | 1057 | 6.188 | 0.2773 | Yes | ||
| 24 | SMTN | 1114 | 6.053 | 0.2854 | Yes | ||
| 25 | DNMT3A | 1150 | 5.973 | 0.2944 | Yes | ||
| 26 | RUNX1 | 1167 | 5.918 | 0.3041 | Yes | ||
| 27 | STAT3 | 1171 | 5.916 | 0.3144 | Yes | ||
| 28 | XPR1 | 1172 | 5.916 | 0.3249 | Yes | ||
| 29 | PLAG1 | 1336 | 5.534 | 0.3272 | Yes | ||
| 30 | UBE2R2 | 1356 | 5.488 | 0.3360 | Yes | ||
| 31 | THAP11 | 1370 | 5.448 | 0.3451 | Yes | ||
| 32 | ZDHHC14 | 1496 | 5.217 | 0.3486 | Yes | ||
| 33 | SEC24C | 1616 | 5.021 | 0.3520 | Yes | ||
| 34 | REV3L | 1675 | 4.930 | 0.3580 | Yes | ||
| 35 | PBX2 | 1696 | 4.901 | 0.3658 | Yes | ||
| 36 | PHF6 | 1864 | 4.663 | 0.3664 | Yes | ||
| 37 | OBFC1 | 2123 | 4.312 | 0.3622 | Yes | ||
| 38 | RTN4 | 2205 | 4.221 | 0.3659 | Yes | ||
| 39 | NLK | 2430 | 3.973 | 0.3626 | Yes | ||
| 40 | PCTK1 | 2434 | 3.967 | 0.3695 | Yes | ||
| 41 | JUP | 2455 | 3.944 | 0.3756 | Yes | ||
| 42 | PLAGL2 | 2501 | 3.889 | 0.3804 | Yes | ||
| 43 | CHD2 | 2595 | 3.816 | 0.3829 | Yes | ||
| 44 | EIF2C3 | 2670 | 3.733 | 0.3861 | Yes | ||
| 45 | TOB1 | 2714 | 3.696 | 0.3906 | Yes | ||
| 46 | PPP2R5E | 2818 | 3.598 | 0.3923 | Yes | ||
| 47 | CDK6 | 2833 | 3.585 | 0.3980 | Yes | ||
| 48 | ANKRD17 | 2918 | 3.502 | 0.4003 | Yes | ||
| 49 | GSK3B | 2980 | 3.451 | 0.4036 | Yes | ||
| 50 | CALM3 | 3006 | 3.431 | 0.4085 | Yes | ||
| 51 | HNRPL | 3027 | 3.413 | 0.4136 | Yes | ||
| 52 | PURA | 3107 | 3.339 | 0.4159 | Yes | ||
| 53 | HDAC3 | 3209 | 3.253 | 0.4170 | Yes | ||
| 54 | DNAJB5 | 3210 | 3.252 | 0.4228 | Yes | ||
| 55 | TRERF1 | 3248 | 3.224 | 0.4268 | Yes | ||
| 56 | KDELR1 | 3667 | 2.898 | 0.4127 | No | ||
| 57 | MAP1A | 3878 | 2.742 | 0.4079 | No | ||
| 58 | CDH3 | 3897 | 2.728 | 0.4119 | No | ||
| 59 | IGSF4D | 3994 | 2.651 | 0.4122 | No | ||
| 60 | EP300 | 4765 | 2.162 | 0.3807 | No | ||
| 61 | SP3 | 4766 | 2.161 | 0.3845 | No | ||
| 62 | GRIN2B | 5286 | 1.882 | 0.3640 | No | ||
| 63 | SAMD8 | 5367 | 1.849 | 0.3636 | No | ||
| 64 | MPL | 5640 | 1.705 | 0.3541 | No | ||
| 65 | NKIRAS2 | 5648 | 1.702 | 0.3568 | No | ||
| 66 | RHOG | 5682 | 1.691 | 0.3583 | No | ||
| 67 | SPAG7 | 6637 | 1.237 | 0.3166 | No | ||
| 68 | FLT1 | 6797 | 1.175 | 0.3114 | No | ||
| 69 | NLGN2 | 7107 | 1.035 | 0.2990 | No | ||
| 70 | SLC12A5 | 7123 | 1.028 | 0.3002 | No | ||
| 71 | TMOD4 | 7228 | 0.984 | 0.2971 | No | ||
| 72 | PRKCH | 7283 | 0.959 | 0.2964 | No | ||
| 73 | NPR2 | 7309 | 0.946 | 0.2969 | No | ||
| 74 | HHEX | 7379 | 0.918 | 0.2953 | No | ||
| 75 | PODN | 7446 | 0.889 | 0.2939 | No | ||
| 76 | CDX1 | 7560 | 0.845 | 0.2902 | No | ||
| 77 | LAMA1 | 7564 | 0.843 | 0.2915 | No | ||
| 78 | MIP | 7637 | 0.813 | 0.2897 | No | ||
| 79 | KLF16 | 7680 | 0.792 | 0.2891 | No | ||
| 80 | ODF3 | 7791 | 0.745 | 0.2854 | No | ||
| 81 | ACCN3 | 7839 | 0.722 | 0.2845 | No | ||
| 82 | OTX2 | 7936 | 0.690 | 0.2813 | No | ||
| 83 | MAPRE3 | 7967 | 0.675 | 0.2811 | No | ||
| 84 | PIM1 | 8068 | 0.635 | 0.2777 | No | ||
| 85 | ACADSB | 8435 | 0.483 | 0.2617 | No | ||
| 86 | SPA17 | 8474 | 0.464 | 0.2608 | No | ||
| 87 | ITPKA | 8518 | 0.445 | 0.2596 | No | ||
| 88 | ITGB8 | 9268 | 0.123 | 0.2254 | No | ||
| 89 | HOXB8 | 9566 | -0.026 | 0.2118 | No | ||
| 90 | BMP2 | 9579 | -0.030 | 0.2113 | No | ||
| 91 | ADAMTS7 | 9594 | -0.039 | 0.2107 | No | ||
| 92 | WNT10A | 9640 | -0.061 | 0.2088 | No | ||
| 93 | EFNB3 | 9743 | -0.106 | 0.2043 | No | ||
| 94 | HOXB6 | 9880 | -0.167 | 0.1983 | No | ||
| 95 | CACNA1G | 9943 | -0.194 | 0.1958 | No | ||
| 96 | KIF9 | 9986 | -0.214 | 0.1943 | No | ||
| 97 | DUSP14 | 10085 | -0.257 | 0.1902 | No | ||
| 98 | STAG2 | 10386 | -0.368 | 0.1771 | No | ||
| 99 | GADD45G | 10557 | -0.424 | 0.1701 | No | ||
| 100 | EED | 10573 | -0.430 | 0.1701 | No | ||
| 101 | NR1D1 | 10596 | -0.438 | 0.1699 | No | ||
| 102 | CASKIN2 | 10778 | -0.506 | 0.1625 | No | ||
| 103 | LHX2 | 10846 | -0.527 | 0.1603 | No | ||
| 104 | KCNJ11 | 11207 | -0.658 | 0.1450 | No | ||
| 105 | HOXC4 | 11324 | -0.689 | 0.1408 | No | ||
| 106 | ASB7 | 11688 | -0.820 | 0.1256 | No | ||
| 107 | FZD2 | 11713 | -0.830 | 0.1260 | No | ||
| 108 | FEV | 11900 | -0.889 | 0.1190 | No | ||
| 109 | BCL3 | 12040 | -0.931 | 0.1143 | No | ||
| 110 | BAI2 | 12283 | -1.002 | 0.1049 | No | ||
| 111 | KLF14 | 12304 | -1.008 | 0.1058 | No | ||
| 112 | CHST11 | 12496 | -1.067 | 0.0989 | No | ||
| 113 | ADAM11 | 12585 | -1.091 | 0.0968 | No | ||
| 114 | PRDM8 | 12593 | -1.092 | 0.0984 | No | ||
| 115 | TPBG | 12741 | -1.137 | 0.0937 | No | ||
| 116 | KCNIP2 | 12779 | -1.150 | 0.0940 | No | ||
| 117 | PHOX2A | 12935 | -1.195 | 0.0890 | No | ||
| 118 | IRS1 | 13166 | -1.265 | 0.0807 | No | ||
| 119 | LIN28 | 13412 | -1.335 | 0.0718 | No | ||
| 120 | BCL6B | 13713 | -1.431 | 0.0605 | No | ||
| 121 | ROM1 | 13745 | -1.438 | 0.0617 | No | ||
| 122 | SCN5A | 13788 | -1.447 | 0.0623 | No | ||
| 123 | GRM7 | 13829 | -1.459 | 0.0630 | No | ||
| 124 | NOL4 | 13922 | -1.484 | 0.0614 | No | ||
| 125 | HSD3B7 | 13979 | -1.500 | 0.0615 | No | ||
| 126 | EPHB1 | 13989 | -1.503 | 0.0638 | No | ||
| 127 | EIF4G1 | 14061 | -1.526 | 0.0632 | No | ||
| 128 | ASCL1 | 14234 | -1.578 | 0.0581 | No | ||
| 129 | CREB3L1 | 14358 | -1.611 | 0.0553 | No | ||
| 130 | CLSTN3 | 14513 | -1.651 | 0.0511 | No | ||
| 131 | SLC6A14 | 14629 | -1.690 | 0.0488 | No | ||
| 132 | PPP2R5D | 14684 | -1.704 | 0.0494 | No | ||
| 133 | BARHL1 | 14757 | -1.729 | 0.0491 | No | ||
| 134 | ARHGAP6 | 14933 | -1.779 | 0.0442 | No | ||
| 135 | GLRA1 | 15559 | -1.986 | 0.0190 | No | ||
| 136 | GRIN2A | 15580 | -1.993 | 0.0216 | No | ||
| 137 | ADAMTS3 | 15755 | -2.048 | 0.0173 | No | ||
| 138 | NR2E1 | 15820 | -2.069 | 0.0180 | No | ||
| 139 | MAGED2 | 15954 | -2.109 | 0.0156 | No | ||
| 140 | CNTN2 | 16016 | -2.126 | 0.0166 | No | ||
| 141 | PRKAG1 | 16085 | -2.147 | 0.0172 | No | ||
| 142 | PPARD | 16135 | -2.163 | 0.0188 | No | ||
| 143 | CPEB3 | 16628 | -2.332 | 0.0003 | No | ||
| 144 | FUNDC1 | 16980 | -2.460 | -0.0114 | No | ||
| 145 | BDNF | 17040 | -2.479 | -0.0097 | No | ||
| 146 | EMX2 | 17217 | -2.542 | -0.0133 | No | ||
| 147 | KCMF1 | 17249 | -2.553 | -0.0102 | No | ||
| 148 | USP2 | 17454 | -2.632 | -0.0150 | No | ||
| 149 | FOXP4 | 17581 | -2.675 | -0.0160 | No | ||
| 150 | AGER | 17698 | -2.726 | -0.0165 | No | ||
| 151 | ZFX | 17937 | -2.827 | -0.0225 | No | ||
| 152 | PHF15 | 18005 | -2.856 | -0.0205 | No | ||
| 153 | BCL7A | 18062 | -2.882 | -0.0180 | No | ||
| 154 | NFATC4 | 18154 | -2.918 | -0.0170 | No | ||
| 155 | WBP5 | 18258 | -2.958 | -0.0165 | No | ||
| 156 | XRN2 | 18427 | -3.037 | -0.0188 | No | ||
| 157 | ALDOC | 18428 | -3.037 | -0.0134 | No | ||
| 158 | RAB34 | 18478 | -3.062 | -0.0103 | No | ||
| 159 | FAF1 | 18567 | -3.102 | -0.0088 | No | ||
| 160 | CNTFR | 19002 | -3.356 | -0.0228 | No | ||
| 161 | CHCHD7 | 19061 | -3.388 | -0.0195 | No | ||
| 162 | PFKFB3 | 19165 | -3.460 | -0.0181 | No | ||
| 163 | PVRL1 | 19191 | -3.477 | -0.0131 | No | ||
| 164 | MAPK10 | 19290 | -3.532 | -0.0114 | No | ||
| 165 | TNNI2 | 19472 | -3.664 | -0.0132 | No | ||
| 166 | MORF4L2 | 19680 | -3.832 | -0.0159 | No | ||
| 167 | H1F0 | 19790 | -3.935 | -0.0140 | No | ||
| 168 | NEO1 | 20318 | -4.470 | -0.0303 | No | ||
| 169 | FUT8 | 20325 | -4.480 | -0.0226 | No | ||
| 170 | RBBP7 | 20378 | -4.547 | -0.0170 | No | ||
| 171 | PABPC1 | 20409 | -4.595 | -0.0102 | No | ||
| 172 | LMO4 | 20804 | -5.199 | -0.0191 | No | ||
| 173 | LAMP2 | 20981 | -5.454 | -0.0176 | No | ||
| 174 | MIB1 | 21173 | -5.895 | -0.0159 | No | ||
| 175 | ATP5G1 | 21410 | -6.613 | -0.0150 | No | ||
| 176 | PLTP | 21614 | -7.424 | -0.0112 | No | ||
| 177 | BMP7 | 21933 | -14.970 | 0.0006 | No |
