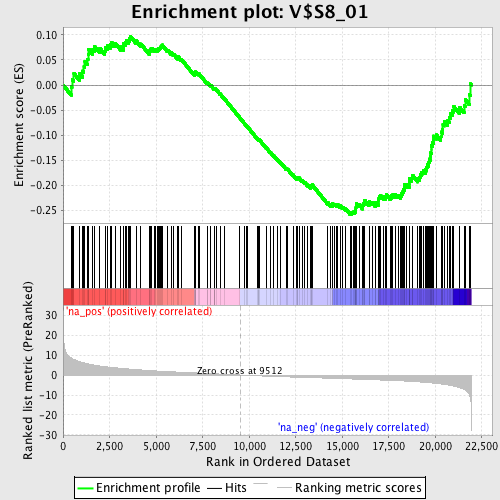
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$S8_01 |
| Enrichment Score (ES) | -0.2575529 |
| Normalized Enrichment Score (NES) | -1.2717725 |
| Nominal p-value | 0.043209877 |
| FDR q-value | 0.3752714 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DEDD | 437 | 8.487 | -0.0028 | No | ||
| 2 | ARID3B | 479 | 8.275 | 0.0121 | No | ||
| 3 | PRKACA | 580 | 7.756 | 0.0232 | No | ||
| 4 | POGZ | 876 | 6.646 | 0.0232 | No | ||
| 5 | FOXP1 | 1045 | 6.223 | 0.0281 | No | ||
| 6 | CYFIP2 | 1121 | 6.046 | 0.0369 | No | ||
| 7 | STAT3 | 1171 | 5.916 | 0.0467 | No | ||
| 8 | ARNTL | 1310 | 5.592 | 0.0517 | No | ||
| 9 | CUGBP2 | 1341 | 5.526 | 0.0616 | No | ||
| 10 | MLLT10 | 1364 | 5.457 | 0.0716 | No | ||
| 11 | TLE4 | 1595 | 5.061 | 0.0713 | No | ||
| 12 | TESK2 | 1692 | 4.905 | 0.0769 | No | ||
| 13 | AP1GBP1 | 1972 | 4.501 | 0.0732 | No | ||
| 14 | RNF122 | 2250 | 4.171 | 0.0690 | No | ||
| 15 | PHC2 | 2286 | 4.125 | 0.0757 | No | ||
| 16 | OGT | 2385 | 4.030 | 0.0794 | No | ||
| 17 | B3GALT1 | 2528 | 3.866 | 0.0807 | No | ||
| 18 | MBNL1 | 2609 | 3.802 | 0.0848 | No | ||
| 19 | XYLT2 | 2801 | 3.608 | 0.0833 | No | ||
| 20 | PURA | 3107 | 3.339 | 0.0761 | No | ||
| 21 | NRXN1 | 3253 | 3.223 | 0.0760 | No | ||
| 22 | BMP1 | 3260 | 3.219 | 0.0822 | No | ||
| 23 | CTNND1 | 3343 | 3.144 | 0.0849 | No | ||
| 24 | TBX6 | 3402 | 3.103 | 0.0885 | No | ||
| 25 | PPP1R16A | 3532 | 2.991 | 0.0886 | No | ||
| 26 | RFX3 | 3564 | 2.971 | 0.0933 | No | ||
| 27 | MYST4 | 3603 | 2.947 | 0.0975 | No | ||
| 28 | SLC25A25 | 3920 | 2.705 | 0.0885 | No | ||
| 29 | TYR | 4175 | 2.531 | 0.0819 | No | ||
| 30 | ZNF148 | 4643 | 2.231 | 0.0650 | No | ||
| 31 | OPRM1 | 4659 | 2.223 | 0.0688 | No | ||
| 32 | GRM5 | 4691 | 2.204 | 0.0719 | No | ||
| 33 | KCNK2 | 4761 | 2.164 | 0.0731 | No | ||
| 34 | SSH2 | 4916 | 2.088 | 0.0703 | No | ||
| 35 | DIO2 | 4987 | 2.045 | 0.0712 | No | ||
| 36 | NRP1 | 5071 | 1.990 | 0.0715 | No | ||
| 37 | DIXDC1 | 5128 | 1.963 | 0.0729 | No | ||
| 38 | SLC26A7 | 5194 | 1.932 | 0.0738 | No | ||
| 39 | RORB | 5234 | 1.911 | 0.0759 | No | ||
| 40 | CNTNAP4 | 5272 | 1.891 | 0.0780 | No | ||
| 41 | UVRAG | 5314 | 1.869 | 0.0799 | No | ||
| 42 | DSPP | 5611 | 1.721 | 0.0698 | No | ||
| 43 | BTBD3 | 5799 | 1.637 | 0.0646 | No | ||
| 44 | CCNJ | 5936 | 1.578 | 0.0615 | No | ||
| 45 | GBX2 | 6151 | 1.474 | 0.0547 | No | ||
| 46 | PAQR9 | 6175 | 1.463 | 0.0566 | No | ||
| 47 | JPH3 | 6346 | 1.384 | 0.0516 | No | ||
| 48 | PRSS12 | 7053 | 1.058 | 0.0213 | No | ||
| 49 | MS4A1 | 7055 | 1.058 | 0.0234 | No | ||
| 50 | LRRC4 | 7086 | 1.044 | 0.0242 | No | ||
| 51 | NLGN2 | 7107 | 1.035 | 0.0254 | No | ||
| 52 | MITF | 7130 | 1.025 | 0.0264 | No | ||
| 53 | STC1 | 7271 | 0.963 | 0.0219 | No | ||
| 54 | HRK | 7307 | 0.948 | 0.0223 | No | ||
| 55 | LRRTM1 | 7742 | 0.763 | 0.0039 | No | ||
| 56 | ITPR3 | 7763 | 0.754 | 0.0045 | No | ||
| 57 | PDZRN4 | 7902 | 0.702 | -0.0004 | No | ||
| 58 | OTX2 | 7936 | 0.690 | -0.0005 | No | ||
| 59 | USH3A | 8122 | 0.610 | -0.0078 | No | ||
| 60 | PRKAB1 | 8148 | 0.596 | -0.0077 | No | ||
| 61 | IRX4 | 8152 | 0.595 | -0.0067 | No | ||
| 62 | LMO3 | 8247 | 0.558 | -0.0098 | No | ||
| 63 | ACADSB | 8435 | 0.483 | -0.0175 | No | ||
| 64 | SYTL2 | 8653 | 0.387 | -0.0266 | No | ||
| 65 | SLC24A4 | 9453 | 0.035 | -0.0633 | No | ||
| 66 | EFNB3 | 9743 | -0.106 | -0.0763 | No | ||
| 67 | TCAP | 9841 | -0.154 | -0.0805 | No | ||
| 68 | HOXB6 | 9880 | -0.167 | -0.0819 | No | ||
| 69 | PIPOX | 10447 | -0.388 | -0.1071 | No | ||
| 70 | IRX6 | 10512 | -0.408 | -0.1092 | No | ||
| 71 | LEMD1 | 10519 | -0.412 | -0.1086 | No | ||
| 72 | CGA | 10539 | -0.419 | -0.1087 | No | ||
| 73 | SALL3 | 10912 | -0.553 | -0.1246 | No | ||
| 74 | DEPDC2 | 11144 | -0.636 | -0.1340 | No | ||
| 75 | HOXC4 | 11324 | -0.689 | -0.1408 | No | ||
| 76 | CLDN8 | 11533 | -0.760 | -0.1488 | No | ||
| 77 | ASB7 | 11688 | -0.820 | -0.1542 | No | ||
| 78 | GPRC5C | 12001 | -0.922 | -0.1667 | No | ||
| 79 | SLC6A15 | 12049 | -0.935 | -0.1669 | No | ||
| 80 | LDB2 | 12355 | -1.023 | -0.1789 | No | ||
| 81 | FIGN | 12549 | -1.080 | -0.1855 | No | ||
| 82 | ADAM11 | 12585 | -1.091 | -0.1849 | No | ||
| 83 | PRDM8 | 12593 | -1.092 | -0.1830 | No | ||
| 84 | SCN3A | 12680 | -1.117 | -0.1847 | No | ||
| 85 | SEZ6 | 12840 | -1.168 | -0.1896 | No | ||
| 86 | TRPM8 | 12996 | -1.217 | -0.1943 | No | ||
| 87 | ANK2 | 13146 | -1.259 | -0.1986 | No | ||
| 88 | EBF2 | 13295 | -1.300 | -0.2027 | No | ||
| 89 | WNT8B | 13322 | -1.308 | -0.2013 | No | ||
| 90 | ZFPM2 | 13347 | -1.312 | -0.1997 | No | ||
| 91 | PRG4 | 13375 | -1.324 | -0.1983 | No | ||
| 92 | UBE1C | 14223 | -1.574 | -0.2340 | No | ||
| 93 | GAP43 | 14389 | -1.617 | -0.2383 | No | ||
| 94 | POLE2 | 14478 | -1.640 | -0.2390 | No | ||
| 95 | LRFN5 | 14484 | -1.641 | -0.2359 | No | ||
| 96 | SIX3 | 14583 | -1.675 | -0.2370 | No | ||
| 97 | KCTD15 | 14702 | -1.713 | -0.2389 | No | ||
| 98 | NPAS3 | 14742 | -1.724 | -0.2372 | No | ||
| 99 | DNAJC5B | 14884 | -1.765 | -0.2401 | No | ||
| 100 | SLC24A2 | 15026 | -1.808 | -0.2429 | No | ||
| 101 | HOXA9 | 15182 | -1.861 | -0.2463 | No | ||
| 102 | PHOX2B | 15429 | -1.933 | -0.2536 | Yes | ||
| 103 | PROP1 | 15507 | -1.962 | -0.2532 | Yes | ||
| 104 | GRIN2A | 15580 | -1.993 | -0.2524 | Yes | ||
| 105 | HOXC6 | 15663 | -2.021 | -0.2521 | Yes | ||
| 106 | SH3BGRL2 | 15696 | -2.031 | -0.2494 | Yes | ||
| 107 | EOMES | 15729 | -2.040 | -0.2468 | Yes | ||
| 108 | DCT | 15734 | -2.042 | -0.2428 | Yes | ||
| 109 | ETV1 | 15743 | -2.044 | -0.2390 | Yes | ||
| 110 | ADAMTS3 | 15755 | -2.048 | -0.2354 | Yes | ||
| 111 | CD36 | 15901 | -2.095 | -0.2378 | Yes | ||
| 112 | GCNT2 | 16098 | -2.150 | -0.2424 | Yes | ||
| 113 | HOXA7 | 16112 | -2.156 | -0.2386 | Yes | ||
| 114 | GRPR | 16148 | -2.165 | -0.2359 | Yes | ||
| 115 | RGS8 | 16179 | -2.174 | -0.2328 | Yes | ||
| 116 | CLSTN2 | 16188 | -2.176 | -0.2288 | Yes | ||
| 117 | MTUS1 | 16440 | -2.267 | -0.2357 | Yes | ||
| 118 | TAC1 | 16459 | -2.274 | -0.2319 | Yes | ||
| 119 | HAS2 | 16601 | -2.323 | -0.2337 | Yes | ||
| 120 | EFNA5 | 16783 | -2.387 | -0.2371 | Yes | ||
| 121 | DMD | 16796 | -2.392 | -0.2328 | Yes | ||
| 122 | MAB21L1 | 16941 | -2.445 | -0.2345 | Yes | ||
| 123 | CXCL14 | 16943 | -2.447 | -0.2296 | Yes | ||
| 124 | SULF2 | 16966 | -2.454 | -0.2256 | Yes | ||
| 125 | PTHR1 | 16991 | -2.464 | -0.2217 | Yes | ||
| 126 | CALD1 | 17060 | -2.487 | -0.2197 | Yes | ||
| 127 | MEF2C | 17220 | -2.542 | -0.2219 | Yes | ||
| 128 | HOXD8 | 17337 | -2.592 | -0.2220 | Yes | ||
| 129 | SHOX2 | 17350 | -2.597 | -0.2172 | Yes | ||
| 130 | DDX5 | 17599 | -2.683 | -0.2232 | Yes | ||
| 131 | HEY2 | 17641 | -2.702 | -0.2196 | Yes | ||
| 132 | FST | 17720 | -2.736 | -0.2176 | Yes | ||
| 133 | HESX1 | 17852 | -2.792 | -0.2180 | Yes | ||
| 134 | OSR2 | 17997 | -2.853 | -0.2188 | Yes | ||
| 135 | NFATC4 | 18154 | -2.918 | -0.2200 | Yes | ||
| 136 | SLC39A14 | 18183 | -2.929 | -0.2154 | Yes | ||
| 137 | PRRX1 | 18256 | -2.957 | -0.2127 | Yes | ||
| 138 | BMP5 | 18290 | -2.971 | -0.2081 | Yes | ||
| 139 | ESR2 | 18334 | -2.993 | -0.2040 | Yes | ||
| 140 | DSCAM | 18339 | -2.997 | -0.1981 | Yes | ||
| 141 | ACP6 | 18461 | -3.054 | -0.1975 | Yes | ||
| 142 | MYT1 | 18594 | -3.122 | -0.1972 | Yes | ||
| 143 | ZFHX1B | 18599 | -3.124 | -0.1911 | Yes | ||
| 144 | PARD6G | 18619 | -3.136 | -0.1856 | Yes | ||
| 145 | NPEPL1 | 18746 | -3.209 | -0.1848 | Yes | ||
| 146 | SEMA3A | 18783 | -3.226 | -0.1799 | Yes | ||
| 147 | PDE3B | 19067 | -3.392 | -0.1860 | Yes | ||
| 148 | MAPK8 | 19156 | -3.453 | -0.1831 | Yes | ||
| 149 | EHF | 19185 | -3.474 | -0.1773 | Yes | ||
| 150 | KCNS1 | 19257 | -3.512 | -0.1734 | Yes | ||
| 151 | CNN3 | 19337 | -3.566 | -0.1698 | Yes | ||
| 152 | SOX5 | 19470 | -3.663 | -0.1685 | Yes | ||
| 153 | PIK3R3 | 19528 | -3.708 | -0.1635 | Yes | ||
| 154 | RALGPS2 | 19561 | -3.736 | -0.1574 | Yes | ||
| 155 | GRWD1 | 19651 | -3.808 | -0.1538 | Yes | ||
| 156 | RNF11 | 19674 | -3.827 | -0.1470 | Yes | ||
| 157 | LBX1 | 19742 | -3.891 | -0.1422 | Yes | ||
| 158 | INPP5F | 19756 | -3.902 | -0.1349 | Yes | ||
| 159 | MEIS1 | 19781 | -3.929 | -0.1280 | Yes | ||
| 160 | DLX1 | 19783 | -3.932 | -0.1201 | Yes | ||
| 161 | INPPL1 | 19844 | -3.988 | -0.1147 | Yes | ||
| 162 | HTR2C | 19897 | -4.029 | -0.1089 | Yes | ||
| 163 | OVOL1 | 19911 | -4.043 | -0.1013 | Yes | ||
| 164 | FOSB | 20048 | -4.193 | -0.0990 | Yes | ||
| 165 | SEZ6L | 20304 | -4.450 | -0.1017 | Yes | ||
| 166 | NEO1 | 20318 | -4.470 | -0.0932 | Yes | ||
| 167 | LCP1 | 20395 | -4.572 | -0.0874 | Yes | ||
| 168 | SIX1 | 20410 | -4.597 | -0.0788 | Yes | ||
| 169 | ASF1A | 20470 | -4.688 | -0.0719 | Yes | ||
| 170 | CS | 20672 | -4.961 | -0.0711 | Yes | ||
| 171 | PPAP2B | 20759 | -5.127 | -0.0646 | Yes | ||
| 172 | MCTS1 | 20799 | -5.193 | -0.0559 | Yes | ||
| 173 | FBLN2 | 20909 | -5.332 | -0.0501 | Yes | ||
| 174 | RREB1 | 20991 | -5.466 | -0.0427 | Yes | ||
| 175 | PSMA1 | 21311 | -6.328 | -0.0445 | Yes | ||
| 176 | SOCS5 | 21548 | -7.116 | -0.0409 | Yes | ||
| 177 | PECR | 21603 | -7.342 | -0.0285 | Yes | ||
| 178 | COMMD10 | 21845 | -10.075 | -0.0191 | Yes | ||
| 179 | TRPM1 | 21891 | -11.674 | 0.0026 | Yes |
