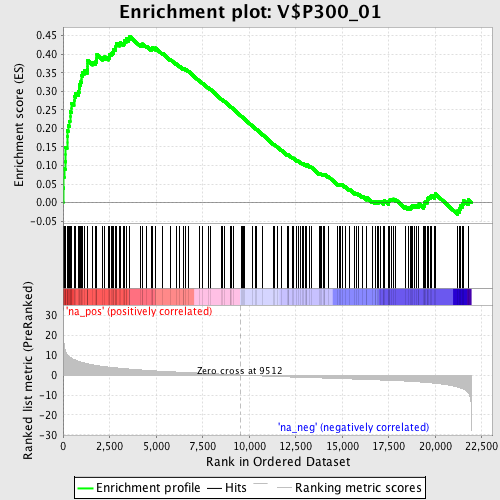
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$P300_01 |
| Enrichment Score (ES) | 0.44862875 |
| Normalized Enrichment Score (NES) | 2.027668 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.008241257 |
| FWER p-Value | 0.012 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EGR3 | 3 | 25.122 | 0.0397 | Yes | ||
| 2 | TNFRSF1A | 22 | 18.882 | 0.0689 | Yes | ||
| 3 | BCL2L1 | 52 | 15.577 | 0.0922 | Yes | ||
| 4 | RORC | 108 | 12.686 | 0.1099 | Yes | ||
| 5 | NFKBIA | 116 | 12.486 | 0.1293 | Yes | ||
| 6 | ZBTB9 | 118 | 12.444 | 0.1490 | Yes | ||
| 7 | WDTC1 | 210 | 10.452 | 0.1615 | Yes | ||
| 8 | PIK3CD | 211 | 10.436 | 0.1780 | Yes | ||
| 9 | ARF6 | 219 | 10.345 | 0.1941 | Yes | ||
| 10 | RGS14 | 269 | 9.747 | 0.2073 | Yes | ||
| 11 | RBMS2 | 329 | 9.268 | 0.2193 | Yes | ||
| 12 | MAP4K2 | 372 | 8.936 | 0.2316 | Yes | ||
| 13 | JUNB | 376 | 8.916 | 0.2456 | Yes | ||
| 14 | PML | 463 | 8.372 | 0.2549 | Yes | ||
| 15 | PDCD10 | 466 | 8.358 | 0.2681 | Yes | ||
| 16 | RARA | 585 | 7.736 | 0.2749 | Yes | ||
| 17 | PPP4C | 591 | 7.711 | 0.2869 | Yes | ||
| 18 | RASGRP2 | 668 | 7.421 | 0.2952 | Yes | ||
| 19 | NFATC1 | 826 | 6.800 | 0.2988 | Yes | ||
| 20 | RFX2 | 859 | 6.698 | 0.3080 | Yes | ||
| 21 | SFRS14 | 884 | 6.627 | 0.3174 | Yes | ||
| 22 | FBS1 | 937 | 6.504 | 0.3253 | Yes | ||
| 23 | PPP3CB | 991 | 6.360 | 0.3330 | Yes | ||
| 24 | JARID2 | 993 | 6.352 | 0.3430 | Yes | ||
| 25 | FOXP1 | 1045 | 6.223 | 0.3505 | Yes | ||
| 26 | PRKCBP1 | 1135 | 6.012 | 0.3560 | Yes | ||
| 27 | ZFYVE26 | 1288 | 5.638 | 0.3580 | Yes | ||
| 28 | BAHD1 | 1315 | 5.575 | 0.3656 | Yes | ||
| 29 | DDAH2 | 1319 | 5.569 | 0.3743 | Yes | ||
| 30 | ENSA | 1323 | 5.567 | 0.3830 | Yes | ||
| 31 | GSN | 1591 | 5.065 | 0.3788 | Yes | ||
| 32 | KCNH3 | 1750 | 4.822 | 0.3792 | Yes | ||
| 33 | SDF2 | 1797 | 4.754 | 0.3846 | Yes | ||
| 34 | RERE | 1811 | 4.736 | 0.3915 | Yes | ||
| 35 | CHD4 | 1814 | 4.734 | 0.3989 | Yes | ||
| 36 | SLC12A4 | 2139 | 4.291 | 0.3909 | Yes | ||
| 37 | MTA2 | 2245 | 4.175 | 0.3927 | Yes | ||
| 38 | NLK | 2430 | 3.973 | 0.3905 | Yes | ||
| 39 | PICALM | 2482 | 3.908 | 0.3944 | Yes | ||
| 40 | EMP1 | 2509 | 3.886 | 0.3994 | Yes | ||
| 41 | PRKACB | 2592 | 3.816 | 0.4017 | Yes | ||
| 42 | ERBB2IP | 2645 | 3.758 | 0.4052 | Yes | ||
| 43 | ACIN1 | 2707 | 3.700 | 0.4083 | Yes | ||
| 44 | SHC1 | 2716 | 3.694 | 0.4138 | Yes | ||
| 45 | CENTD2 | 2822 | 3.596 | 0.4147 | Yes | ||
| 46 | DDIT4 | 2835 | 3.583 | 0.4198 | Yes | ||
| 47 | KCNAB2 | 2843 | 3.574 | 0.4252 | Yes | ||
| 48 | SRPK2 | 2883 | 3.531 | 0.4290 | Yes | ||
| 49 | PCTK2 | 3038 | 3.404 | 0.4273 | Yes | ||
| 50 | BCL11A | 3071 | 3.369 | 0.4312 | Yes | ||
| 51 | MYCL1 | 3237 | 3.234 | 0.4287 | Yes | ||
| 52 | PHC3 | 3276 | 3.201 | 0.4321 | Yes | ||
| 53 | ARHGAP1 | 3297 | 3.177 | 0.4362 | Yes | ||
| 54 | MAP3K4 | 3381 | 3.115 | 0.4373 | Yes | ||
| 55 | SERPINI1 | 3393 | 3.106 | 0.4417 | Yes | ||
| 56 | SUPT6H | 3540 | 2.984 | 0.4398 | Yes | ||
| 57 | DGKA | 3551 | 2.979 | 0.4440 | Yes | ||
| 58 | KIF3B | 3555 | 2.976 | 0.4486 | Yes | ||
| 59 | FURIN | 4152 | 2.546 | 0.4253 | No | ||
| 60 | ZHX2 | 4253 | 2.468 | 0.4246 | No | ||
| 61 | POLD4 | 4275 | 2.455 | 0.4276 | No | ||
| 62 | DMTF1 | 4503 | 2.322 | 0.4208 | No | ||
| 63 | EPN3 | 4767 | 2.161 | 0.4122 | No | ||
| 64 | CRIM1 | 4784 | 2.153 | 0.4148 | No | ||
| 65 | LDOC1 | 4792 | 2.150 | 0.4179 | No | ||
| 66 | SLC9A6 | 4943 | 2.074 | 0.4143 | No | ||
| 67 | DPYSL2 | 4962 | 2.060 | 0.4168 | No | ||
| 68 | BLCAP | 5363 | 1.850 | 0.4013 | No | ||
| 69 | SYN1 | 5776 | 1.649 | 0.3850 | No | ||
| 70 | PUM2 | 6074 | 1.509 | 0.3738 | No | ||
| 71 | TNFRSF12A | 6258 | 1.423 | 0.3676 | No | ||
| 72 | BHLHB3 | 6445 | 1.339 | 0.3612 | No | ||
| 73 | WNT7B | 6494 | 1.315 | 0.3611 | No | ||
| 74 | CAST | 6585 | 1.267 | 0.3590 | No | ||
| 75 | WNT11 | 6729 | 1.203 | 0.3543 | No | ||
| 76 | PET112L | 7325 | 0.939 | 0.3285 | No | ||
| 77 | ZCCHC5 | 7498 | 0.869 | 0.3220 | No | ||
| 78 | AAMP | 7794 | 0.744 | 0.3096 | No | ||
| 79 | CSMD3 | 7920 | 0.696 | 0.3050 | No | ||
| 80 | NXPH4 | 8515 | 0.446 | 0.2784 | No | ||
| 81 | PPP1CB | 8547 | 0.434 | 0.2777 | No | ||
| 82 | FBXL19 | 8665 | 0.384 | 0.2729 | No | ||
| 83 | WNT4 | 8968 | 0.248 | 0.2594 | No | ||
| 84 | FDPS | 9005 | 0.231 | 0.2581 | No | ||
| 85 | WNT3 | 9060 | 0.205 | 0.2560 | No | ||
| 86 | GFRA3 | 9172 | 0.162 | 0.2511 | No | ||
| 87 | GPLD1 | 9600 | -0.045 | 0.2316 | No | ||
| 88 | WNT10A | 9640 | -0.061 | 0.2299 | No | ||
| 89 | VAX1 | 9669 | -0.074 | 0.2287 | No | ||
| 90 | EFNB3 | 9743 | -0.106 | 0.2256 | No | ||
| 91 | PPP1R1B | 10155 | -0.280 | 0.2071 | No | ||
| 92 | PNOC | 10333 | -0.352 | 0.1995 | No | ||
| 93 | STAG2 | 10386 | -0.368 | 0.1977 | No | ||
| 94 | HEBP1 | 10731 | -0.487 | 0.1827 | No | ||
| 95 | HOXC4 | 11324 | -0.689 | 0.1566 | No | ||
| 96 | INHA | 11336 | -0.692 | 0.1572 | No | ||
| 97 | GABRA1 | 11521 | -0.757 | 0.1500 | No | ||
| 98 | FZD5 | 11754 | -0.843 | 0.1406 | No | ||
| 99 | S100A16 | 12070 | -0.940 | 0.1277 | No | ||
| 100 | GJB1 | 12095 | -0.948 | 0.1281 | No | ||
| 101 | ESRRG | 12112 | -0.951 | 0.1289 | No | ||
| 102 | HCN3 | 12344 | -1.020 | 0.1199 | No | ||
| 103 | SLITRK5 | 12368 | -1.029 | 0.1204 | No | ||
| 104 | RHOV | 12566 | -1.086 | 0.1131 | No | ||
| 105 | VMD2L1 | 12623 | -1.104 | 0.1123 | No | ||
| 106 | FGFR1 | 12767 | -1.146 | 0.1075 | No | ||
| 107 | NLGN3 | 12836 | -1.167 | 0.1063 | No | ||
| 108 | RARG | 12909 | -1.189 | 0.1049 | No | ||
| 109 | NPR1 | 13036 | -1.229 | 0.1010 | No | ||
| 110 | TRIM3 | 13092 | -1.243 | 0.1005 | No | ||
| 111 | FSHB | 13096 | -1.244 | 0.1023 | No | ||
| 112 | GATA1 | 13251 | -1.288 | 0.0973 | No | ||
| 113 | ZIC1 | 13330 | -1.309 | 0.0958 | No | ||
| 114 | ZNF23 | 13753 | -1.439 | 0.0787 | No | ||
| 115 | GRM7 | 13829 | -1.459 | 0.0775 | No | ||
| 116 | CACNB2 | 13894 | -1.477 | 0.0769 | No | ||
| 117 | EPHB1 | 13989 | -1.503 | 0.0750 | No | ||
| 118 | SP6 | 14024 | -1.516 | 0.0759 | No | ||
| 119 | NKX6-2 | 14057 | -1.525 | 0.0768 | No | ||
| 120 | ASCL1 | 14234 | -1.578 | 0.0712 | No | ||
| 121 | HMGCS1 | 14755 | -1.729 | 0.0501 | No | ||
| 122 | SREBF2 | 14848 | -1.754 | 0.0486 | No | ||
| 123 | DPP3 | 14917 | -1.775 | 0.0483 | No | ||
| 124 | TUSC3 | 14989 | -1.798 | 0.0479 | No | ||
| 125 | EMCN | 15149 | -1.853 | 0.0436 | No | ||
| 126 | TFEB | 15407 | -1.927 | 0.0348 | No | ||
| 127 | HOXC6 | 15663 | -2.021 | 0.0263 | No | ||
| 128 | HOXA2 | 15771 | -2.054 | 0.0247 | No | ||
| 129 | PTBP2 | 15893 | -2.092 | 0.0224 | No | ||
| 130 | SUMO2 | 16099 | -2.150 | 0.0164 | No | ||
| 131 | STARD13 | 16304 | -2.219 | 0.0106 | No | ||
| 132 | PLP1 | 16325 | -2.224 | 0.0132 | No | ||
| 133 | SLITRK1 | 16616 | -2.328 | 0.0036 | No | ||
| 134 | ZIC3 | 16766 | -2.382 | 0.0005 | No | ||
| 135 | DMD | 16796 | -2.392 | 0.0030 | No | ||
| 136 | FGF12 | 16906 | -2.433 | 0.0018 | No | ||
| 137 | SIRT3 | 16971 | -2.456 | 0.0028 | No | ||
| 138 | GDNF | 17032 | -2.477 | 0.0039 | No | ||
| 139 | LTBR | 17221 | -2.543 | -0.0007 | No | ||
| 140 | NEUROG1 | 17222 | -2.543 | 0.0034 | No | ||
| 141 | DPF3 | 17243 | -2.551 | 0.0065 | No | ||
| 142 | MOAP1 | 17490 | -2.641 | -0.0006 | No | ||
| 143 | GPC3 | 17497 | -2.647 | 0.0033 | No | ||
| 144 | GRK5 | 17526 | -2.656 | 0.0063 | No | ||
| 145 | NR2F1 | 17563 | -2.666 | 0.0088 | No | ||
| 146 | CTBP2 | 17648 | -2.704 | 0.0093 | No | ||
| 147 | PRKAG2 | 17729 | -2.739 | 0.0099 | No | ||
| 148 | ZIC4 | 17876 | -2.800 | 0.0077 | No | ||
| 149 | KCNN3 | 18396 | -3.022 | -0.0114 | No | ||
| 150 | TWIST1 | 18561 | -3.099 | -0.0140 | No | ||
| 151 | CANX | 18647 | -3.149 | -0.0129 | No | ||
| 152 | PPP3CA | 18699 | -3.178 | -0.0102 | No | ||
| 153 | LHX6 | 18749 | -3.210 | -0.0073 | No | ||
| 154 | SYNCRIP | 18869 | -3.273 | -0.0076 | No | ||
| 155 | AKAP12 | 18976 | -3.339 | -0.0072 | No | ||
| 156 | SIX4 | 19099 | -3.414 | -0.0074 | No | ||
| 157 | ELMO1 | 19121 | -3.431 | -0.0029 | No | ||
| 158 | SEMA4C | 19380 | -3.598 | -0.0090 | No | ||
| 159 | CXXC5 | 19406 | -3.615 | -0.0044 | No | ||
| 160 | SMPD3 | 19429 | -3.634 | 0.0003 | No | ||
| 161 | IRAK1 | 19476 | -3.666 | 0.0040 | No | ||
| 162 | IKBKB | 19586 | -3.755 | 0.0050 | No | ||
| 163 | ADAM15 | 19596 | -3.763 | 0.0105 | No | ||
| 164 | GJB2 | 19637 | -3.796 | 0.0147 | No | ||
| 165 | LBX1 | 19742 | -3.891 | 0.0161 | No | ||
| 166 | DLX1 | 19783 | -3.932 | 0.0205 | No | ||
| 167 | ELAVL4 | 19973 | -4.101 | 0.0183 | No | ||
| 168 | EBAG9 | 19984 | -4.115 | 0.0244 | No | ||
| 169 | CAMK2D | 21209 | -6.011 | -0.0223 | No | ||
| 170 | ATP8A2 | 21306 | -6.306 | -0.0167 | No | ||
| 171 | NME1 | 21334 | -6.394 | -0.0077 | No | ||
| 172 | CKS1B | 21442 | -6.729 | -0.0020 | No | ||
| 173 | E2F3 | 21509 | -6.976 | 0.0061 | No | ||
| 174 | PSMD13 | 21775 | -8.833 | 0.0079 | No |
