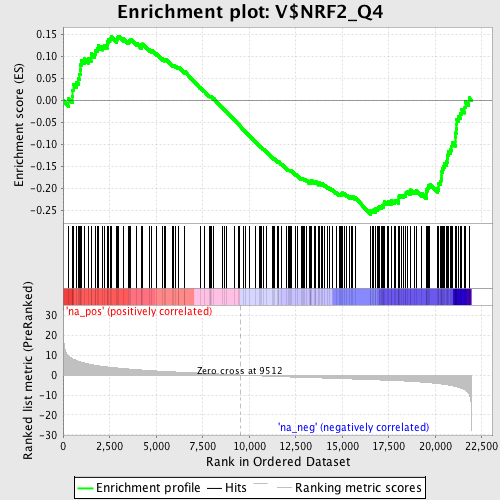
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NRF2_Q4 |
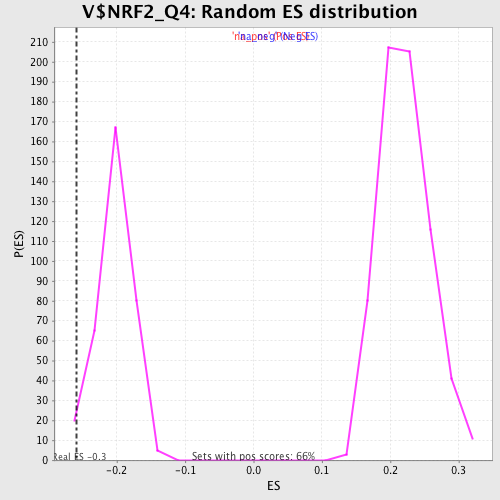
| Enrichment Score (ES) | -0.25864148 |
| Normalized Enrichment Score (NES) | -1.2757827 |
| Nominal p-value | 0.029673591 |
| FDR q-value | 0.39897865 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BAZ2A | 293 | 9.571 | 0.0042 | No | ||
| 2 | RB1CC1 | 492 | 8.182 | 0.0102 | No | ||
| 3 | TUBA1 | 511 | 8.088 | 0.0242 | No | ||
| 4 | RTN3 | 542 | 7.957 | 0.0375 | No | ||
| 5 | PAFAH1B1 | 726 | 7.153 | 0.0423 | No | ||
| 6 | BACH1 | 833 | 6.787 | 0.0499 | No | ||
| 7 | PFN2 | 890 | 6.623 | 0.0596 | No | ||
| 8 | EDG1 | 914 | 6.554 | 0.0706 | No | ||
| 9 | ABCD1 | 940 | 6.494 | 0.0814 | No | ||
| 10 | TBL1X | 973 | 6.407 | 0.0918 | No | ||
| 11 | LAMC1 | 1143 | 5.991 | 0.0950 | No | ||
| 12 | DTX2 | 1354 | 5.496 | 0.0955 | No | ||
| 13 | SQSTM1 | 1504 | 5.209 | 0.0983 | No | ||
| 14 | DDX17 | 1513 | 5.188 | 0.1075 | No | ||
| 15 | ITPKC | 1713 | 4.878 | 0.1073 | No | ||
| 16 | KCNH3 | 1750 | 4.822 | 0.1146 | No | ||
| 17 | SEC24D | 1832 | 4.701 | 0.1195 | No | ||
| 18 | RHOH | 1900 | 4.593 | 0.1249 | No | ||
| 19 | CSNK1G2 | 2114 | 4.319 | 0.1231 | No | ||
| 20 | BRD2 | 2224 | 4.202 | 0.1258 | No | ||
| 21 | CAPN12 | 2362 | 4.050 | 0.1270 | No | ||
| 22 | FBXO30 | 2376 | 4.035 | 0.1338 | No | ||
| 23 | NR5A1 | 2423 | 3.980 | 0.1390 | No | ||
| 24 | SLC38A2 | 2520 | 3.874 | 0.1418 | No | ||
| 25 | H3F3B | 2581 | 3.828 | 0.1461 | No | ||
| 26 | ATP1B1 | 2894 | 3.522 | 0.1382 | No | ||
| 27 | TUBB | 2902 | 3.513 | 0.1444 | No | ||
| 28 | TUBA4 | 2990 | 3.445 | 0.1467 | No | ||
| 29 | BCL9L | 3239 | 3.231 | 0.1413 | No | ||
| 30 | PLEC1 | 3499 | 3.020 | 0.1350 | No | ||
| 31 | BEX2 | 3575 | 2.963 | 0.1370 | No | ||
| 32 | MAST2 | 3643 | 2.912 | 0.1393 | No | ||
| 33 | AKT3 | 3957 | 2.670 | 0.1298 | No | ||
| 34 | PITPNC1 | 4201 | 2.506 | 0.1233 | No | ||
| 35 | RDH5 | 4221 | 2.494 | 0.1270 | No | ||
| 36 | KCNA2 | 4278 | 2.454 | 0.1289 | No | ||
| 37 | PSMD4 | 4636 | 2.236 | 0.1166 | No | ||
| 38 | UBXD2 | 4773 | 2.156 | 0.1144 | No | ||
| 39 | FCHSD1 | 5020 | 2.021 | 0.1068 | No | ||
| 40 | ADCK4 | 5341 | 1.857 | 0.0955 | No | ||
| 41 | KCND1 | 5461 | 1.798 | 0.0934 | No | ||
| 42 | SOST | 5527 | 1.763 | 0.0936 | No | ||
| 43 | IPO13 | 5878 | 1.600 | 0.0805 | No | ||
| 44 | SPTBN4 | 5940 | 1.576 | 0.0806 | No | ||
| 45 | FOXN1 | 6064 | 1.511 | 0.0777 | No | ||
| 46 | PIAS1 | 6189 | 1.455 | 0.0747 | No | ||
| 47 | PSME4 | 6224 | 1.439 | 0.0758 | No | ||
| 48 | MNT | 6512 | 1.303 | 0.0650 | No | ||
| 49 | WNT2 | 6534 | 1.291 | 0.0664 | No | ||
| 50 | F2RL2 | 7375 | 0.920 | 0.0295 | No | ||
| 51 | PRDM1 | 7584 | 0.833 | 0.0215 | No | ||
| 52 | MMRN2 | 7871 | 0.712 | 0.0097 | No | ||
| 53 | PDZRN4 | 7902 | 0.702 | 0.0096 | No | ||
| 54 | CSMD3 | 7920 | 0.696 | 0.0101 | No | ||
| 55 | MAPRE3 | 7967 | 0.675 | 0.0092 | No | ||
| 56 | ABCA2 | 8095 | 0.623 | 0.0045 | No | ||
| 57 | SMPX | 8545 | 0.434 | -0.0153 | No | ||
| 58 | GTF2A1 | 8677 | 0.379 | -0.0206 | No | ||
| 59 | KIF5A | 8767 | 0.333 | -0.0241 | No | ||
| 60 | CRYBA2 | 9200 | 0.153 | -0.0437 | No | ||
| 61 | LAMA3 | 9426 | 0.045 | -0.0539 | No | ||
| 62 | MIA2 | 9498 | 0.008 | -0.0572 | No | ||
| 63 | PSMD2 | 9671 | -0.075 | -0.0649 | No | ||
| 64 | CLDN15 | 9786 | -0.121 | -0.0700 | No | ||
| 65 | UCHL1 | 9999 | -0.220 | -0.0793 | No | ||
| 66 | DUSP13 | 10325 | -0.349 | -0.0936 | No | ||
| 67 | GADD45G | 10557 | -0.424 | -0.1034 | No | ||
| 68 | NR1D1 | 10596 | -0.438 | -0.1044 | No | ||
| 69 | PHLDA2 | 10652 | -0.455 | -0.1060 | No | ||
| 70 | TLL1 | 10780 | -0.507 | -0.1109 | No | ||
| 71 | RGS2 | 10906 | -0.550 | -0.1157 | No | ||
| 72 | CNTN6 | 11230 | -0.664 | -0.1293 | No | ||
| 73 | ABHD4 | 11320 | -0.688 | -0.1321 | No | ||
| 74 | INHA | 11336 | -0.692 | -0.1315 | No | ||
| 75 | DCTN2 | 11539 | -0.762 | -0.1394 | No | ||
| 76 | SNCG | 11564 | -0.774 | -0.1391 | No | ||
| 77 | TNXB | 11599 | -0.784 | -0.1392 | No | ||
| 78 | LRP1B | 11748 | -0.841 | -0.1444 | No | ||
| 79 | BTK | 12025 | -0.929 | -0.1554 | No | ||
| 80 | ESRRG | 12112 | -0.951 | -0.1576 | No | ||
| 81 | PKN3 | 12153 | -0.961 | -0.1577 | No | ||
| 82 | TFPI2 | 12212 | -0.979 | -0.1585 | No | ||
| 83 | NDRG2 | 12273 | -1.000 | -0.1595 | No | ||
| 84 | TBXAS1 | 12507 | -1.069 | -0.1682 | No | ||
| 85 | SYTL1 | 12612 | -1.100 | -0.1709 | No | ||
| 86 | SFXN5 | 12805 | -1.161 | -0.1776 | No | ||
| 87 | GPX2 | 12839 | -1.168 | -0.1770 | No | ||
| 88 | RARG | 12909 | -1.189 | -0.1780 | No | ||
| 89 | FGF9 | 12977 | -1.209 | -0.1788 | No | ||
| 90 | MDFI | 13099 | -1.245 | -0.1821 | No | ||
| 91 | NUDT10 | 13248 | -1.287 | -0.1865 | No | ||
| 92 | NUDT11 | 13279 | -1.295 | -0.1855 | No | ||
| 93 | SYNPO | 13281 | -1.296 | -0.1832 | No | ||
| 94 | MAP2K1 | 13318 | -1.307 | -0.1824 | No | ||
| 95 | ZIC1 | 13330 | -1.309 | -0.1805 | No | ||
| 96 | NOG | 13521 | -1.371 | -0.1867 | No | ||
| 97 | PPP2R2C | 13559 | -1.382 | -0.1858 | No | ||
| 98 | IL6 | 13580 | -1.388 | -0.1842 | No | ||
| 99 | ROM1 | 13745 | -1.438 | -0.1891 | No | ||
| 100 | SLC26A1 | 13754 | -1.439 | -0.1868 | No | ||
| 101 | CACNB2 | 13894 | -1.477 | -0.1905 | No | ||
| 102 | FGF11 | 13918 | -1.484 | -0.1888 | No | ||
| 103 | EIF4G1 | 14061 | -1.526 | -0.1925 | No | ||
| 104 | MCF2 | 14232 | -1.576 | -0.1974 | No | ||
| 105 | HSPG2 | 14324 | -1.603 | -0.1986 | No | ||
| 106 | PDE4D | 14460 | -1.636 | -0.2018 | No | ||
| 107 | TFEC | 14662 | -1.701 | -0.2079 | No | ||
| 108 | OGG1 | 14843 | -1.753 | -0.2130 | No | ||
| 109 | PER2 | 14911 | -1.773 | -0.2128 | No | ||
| 110 | NOTCH4 | 14957 | -1.787 | -0.2116 | No | ||
| 111 | RABEP1 | 14987 | -1.796 | -0.2096 | No | ||
| 112 | RAB6A | 15095 | -1.832 | -0.2111 | No | ||
| 113 | MMP19 | 15244 | -1.880 | -0.2145 | No | ||
| 114 | PPP2R2B | 15396 | -1.924 | -0.2178 | No | ||
| 115 | FGF1 | 15516 | -1.965 | -0.2197 | No | ||
| 116 | FLNC | 15574 | -1.990 | -0.2186 | No | ||
| 117 | SH3BGRL2 | 15696 | -2.031 | -0.2205 | No | ||
| 118 | LGI3 | 16528 | -2.299 | -0.2544 | Yes | ||
| 119 | CRYGS | 16537 | -2.303 | -0.2505 | Yes | ||
| 120 | PPP1R9B | 16637 | -2.334 | -0.2508 | Yes | ||
| 121 | ATP6V0A1 | 16680 | -2.350 | -0.2484 | Yes | ||
| 122 | DMD | 16796 | -2.392 | -0.2492 | Yes | ||
| 123 | ADCYAP1 | 16799 | -2.394 | -0.2449 | Yes | ||
| 124 | FGF12 | 16906 | -2.433 | -0.2453 | Yes | ||
| 125 | APBA1 | 16935 | -2.444 | -0.2421 | Yes | ||
| 126 | CAPN6 | 17007 | -2.468 | -0.2408 | Yes | ||
| 127 | CD44 | 17112 | -2.503 | -0.2410 | Yes | ||
| 128 | EEF1A2 | 17157 | -2.518 | -0.2384 | Yes | ||
| 129 | USP14 | 17240 | -2.550 | -0.2374 | Yes | ||
| 130 | TXNRD1 | 17241 | -2.551 | -0.2327 | Yes | ||
| 131 | ENO1 | 17283 | -2.566 | -0.2299 | Yes | ||
| 132 | GAS2L1 | 17418 | -2.619 | -0.2312 | Yes | ||
| 133 | CDH23 | 17474 | -2.638 | -0.2289 | Yes | ||
| 134 | PRX | 17623 | -2.691 | -0.2307 | Yes | ||
| 135 | NRXN2 | 17628 | -2.692 | -0.2259 | Yes | ||
| 136 | ITM2B | 17789 | -2.768 | -0.2282 | Yes | ||
| 137 | ZIC4 | 17876 | -2.800 | -0.2270 | Yes | ||
| 138 | APBA2BP | 18016 | -2.862 | -0.2281 | Yes | ||
| 139 | TREX1 | 18043 | -2.873 | -0.2240 | Yes | ||
| 140 | IDS | 18046 | -2.875 | -0.2188 | Yes | ||
| 141 | DRP2 | 18080 | -2.890 | -0.2150 | Yes | ||
| 142 | LOXL4 | 18198 | -2.935 | -0.2149 | Yes | ||
| 143 | TGIF | 18307 | -2.979 | -0.2144 | Yes | ||
| 144 | MGST1 | 18403 | -3.025 | -0.2132 | Yes | ||
| 145 | SYT2 | 18423 | -3.035 | -0.2085 | Yes | ||
| 146 | SLC16A6 | 18482 | -3.064 | -0.2055 | Yes | ||
| 147 | CANX | 18647 | -3.149 | -0.2072 | Yes | ||
| 148 | TTC1 | 18680 | -3.166 | -0.2028 | Yes | ||
| 149 | PSMA3 | 18882 | -3.284 | -0.2060 | Yes | ||
| 150 | MASP1 | 18974 | -3.339 | -0.2041 | Yes | ||
| 151 | SKP1A | 19269 | -3.518 | -0.2111 | Yes | ||
| 152 | WBP4 | 19517 | -3.702 | -0.2156 | Yes | ||
| 153 | CPNE8 | 19521 | -3.705 | -0.2089 | Yes | ||
| 154 | ALS2 | 19539 | -3.713 | -0.2029 | Yes | ||
| 155 | ADAM15 | 19596 | -3.763 | -0.1985 | Yes | ||
| 156 | PSMC5 | 19623 | -3.785 | -0.1927 | Yes | ||
| 157 | LIMK1 | 19712 | -3.863 | -0.1896 | Yes | ||
| 158 | MRPL32 | 20116 | -4.259 | -0.2003 | Yes | ||
| 159 | BNIP3 | 20187 | -4.336 | -0.1955 | Yes | ||
| 160 | ABCB6 | 20189 | -4.339 | -0.1876 | Yes | ||
| 161 | ELA1 | 20278 | -4.427 | -0.1835 | Yes | ||
| 162 | USP13 | 20306 | -4.453 | -0.1765 | Yes | ||
| 163 | TIAL1 | 20346 | -4.505 | -0.1700 | Yes | ||
| 164 | PSMD1 | 20350 | -4.509 | -0.1618 | Yes | ||
| 165 | RBBP7 | 20378 | -4.547 | -0.1547 | Yes | ||
| 166 | BLVRB | 20431 | -4.633 | -0.1485 | Yes | ||
| 167 | TOMM70A | 20510 | -4.734 | -0.1434 | Yes | ||
| 168 | YWHAG | 20590 | -4.829 | -0.1381 | Yes | ||
| 169 | LRP1 | 20637 | -4.891 | -0.1312 | Yes | ||
| 170 | BLMH | 20668 | -4.955 | -0.1234 | Yes | ||
| 171 | SLC11A1 | 20705 | -5.033 | -0.1158 | Yes | ||
| 172 | LMO4 | 20804 | -5.199 | -0.1107 | Yes | ||
| 173 | LRRFIP2 | 20884 | -5.301 | -0.1046 | Yes | ||
| 174 | PSMD12 | 20898 | -5.320 | -0.0954 | Yes | ||
| 175 | HOXA3 | 21069 | -5.643 | -0.0928 | Yes | ||
| 176 | ALDOA | 21071 | -5.645 | -0.0824 | Yes | ||
| 177 | XPOT | 21103 | -5.712 | -0.0733 | Yes | ||
| 178 | ETV5 | 21115 | -5.739 | -0.0633 | Yes | ||
| 179 | UBE2L3 | 21148 | -5.829 | -0.0540 | Yes | ||
| 180 | RAN | 21154 | -5.842 | -0.0434 | Yes | ||
| 181 | FTSJ3 | 21247 | -6.128 | -0.0364 | Yes | ||
| 182 | PSMD7 | 21338 | -6.407 | -0.0287 | Yes | ||
| 183 | PSMA2 | 21396 | -6.560 | -0.0192 | Yes | ||
| 184 | BAG2 | 21589 | -7.273 | -0.0146 | Yes | ||
| 185 | EPHB2 | 21625 | -7.511 | -0.0024 | Yes | ||
| 186 | ARHGAP8 | 21809 | -9.307 | 0.0063 | Yes |