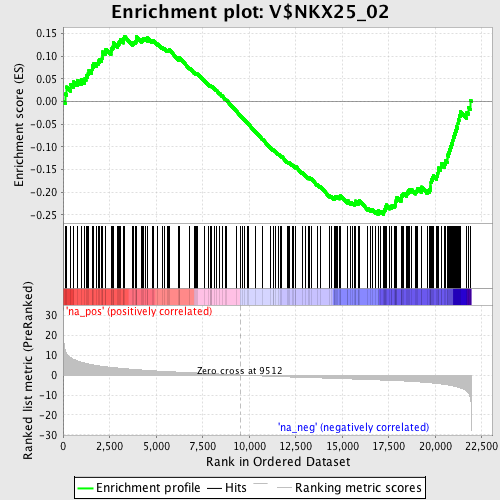
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NKX25_02 |
| Enrichment Score (ES) | -0.24925923 |
| Normalized Enrichment Score (NES) | -1.2251704 |
| Nominal p-value | 0.08256881 |
| FDR q-value | 0.43024296 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SCML4 | 111 | 12.656 | 0.0170 | No | ||
| 2 | TCF12 | 187 | 10.757 | 0.0323 | No | ||
| 3 | CHES1 | 390 | 8.863 | 0.0385 | No | ||
| 4 | MSI2 | 575 | 7.805 | 0.0436 | No | ||
| 5 | LNPEP | 757 | 7.032 | 0.0476 | No | ||
| 6 | TBL1X | 973 | 6.407 | 0.0488 | No | ||
| 7 | STAT3 | 1171 | 5.916 | 0.0501 | No | ||
| 8 | VGLL4 | 1232 | 5.785 | 0.0574 | No | ||
| 9 | PLAG1 | 1336 | 5.534 | 0.0624 | No | ||
| 10 | FLNA | 1385 | 5.421 | 0.0696 | No | ||
| 11 | GABARAPL2 | 1552 | 5.124 | 0.0709 | No | ||
| 12 | FBXW11 | 1563 | 5.105 | 0.0793 | No | ||
| 13 | TRPM7 | 1652 | 4.963 | 0.0840 | No | ||
| 14 | EGLN1 | 1819 | 4.720 | 0.0846 | No | ||
| 15 | DFNB31 | 1883 | 4.619 | 0.0897 | No | ||
| 16 | AP1GBP1 | 1972 | 4.501 | 0.0935 | No | ||
| 17 | EIF4E | 2088 | 4.365 | 0.0959 | No | ||
| 18 | ATF7 | 2112 | 4.331 | 0.1023 | No | ||
| 19 | USP40 | 2118 | 4.314 | 0.1096 | No | ||
| 20 | CFL2 | 2263 | 4.155 | 0.1103 | No | ||
| 21 | RGS12 | 2295 | 4.117 | 0.1160 | No | ||
| 22 | KLF12 | 2575 | 3.833 | 0.1099 | No | ||
| 23 | H3F3B | 2581 | 3.828 | 0.1163 | No | ||
| 24 | CNNM2 | 2635 | 3.772 | 0.1205 | No | ||
| 25 | DCX | 2692 | 3.710 | 0.1244 | No | ||
| 26 | DNAJB4 | 2693 | 3.709 | 0.1308 | No | ||
| 27 | HS3ST2 | 2945 | 3.476 | 0.1254 | No | ||
| 28 | AP1S2 | 2987 | 3.447 | 0.1295 | No | ||
| 29 | PTPRC | 3046 | 3.398 | 0.1327 | No | ||
| 30 | NSD1 | 3067 | 3.376 | 0.1377 | No | ||
| 31 | NRXN1 | 3253 | 3.223 | 0.1348 | No | ||
| 32 | ROBO3 | 3261 | 3.219 | 0.1401 | No | ||
| 33 | RNF39 | 3288 | 3.185 | 0.1445 | No | ||
| 34 | INHBA | 3744 | 2.844 | 0.1285 | No | ||
| 35 | CTLA4 | 3798 | 2.805 | 0.1310 | No | ||
| 36 | CTSK | 3876 | 2.742 | 0.1322 | No | ||
| 37 | SLC25A25 | 3920 | 2.705 | 0.1350 | No | ||
| 38 | EDEM1 | 3938 | 2.692 | 0.1389 | No | ||
| 39 | RAB5B | 3942 | 2.683 | 0.1434 | No | ||
| 40 | WDR33 | 4222 | 2.494 | 0.1349 | No | ||
| 41 | VLDLR | 4261 | 2.464 | 0.1375 | No | ||
| 42 | FBXW7 | 4301 | 2.440 | 0.1400 | No | ||
| 43 | ATP11C | 4425 | 2.367 | 0.1384 | No | ||
| 44 | AQP3 | 4540 | 2.301 | 0.1372 | No | ||
| 45 | NR4A3 | 4550 | 2.289 | 0.1408 | No | ||
| 46 | PDE7A | 4800 | 2.147 | 0.1331 | No | ||
| 47 | RNF111 | 4856 | 2.118 | 0.1342 | No | ||
| 48 | LPHN2 | 5084 | 1.984 | 0.1273 | No | ||
| 49 | SEMA5B | 5326 | 1.864 | 0.1194 | No | ||
| 50 | VIP | 5441 | 1.813 | 0.1174 | No | ||
| 51 | DSPP | 5611 | 1.721 | 0.1126 | No | ||
| 52 | OTP | 5656 | 1.699 | 0.1135 | No | ||
| 53 | PPM1E | 5694 | 1.688 | 0.1148 | No | ||
| 54 | CNTN4 | 6216 | 1.441 | 0.0933 | No | ||
| 55 | MAP3K7IP2 | 6233 | 1.435 | 0.0951 | No | ||
| 56 | HMGN2 | 6245 | 1.428 | 0.0971 | No | ||
| 57 | FLT1 | 6797 | 1.175 | 0.0738 | No | ||
| 58 | MS4A1 | 7055 | 1.058 | 0.0639 | No | ||
| 59 | MAP2K5 | 7135 | 1.023 | 0.0620 | No | ||
| 60 | LRP2 | 7184 | 1.004 | 0.0615 | No | ||
| 61 | ABLIM2 | 7242 | 0.978 | 0.0606 | No | ||
| 62 | KCNJ3 | 7570 | 0.841 | 0.0471 | No | ||
| 63 | BBOX1 | 7817 | 0.732 | 0.0370 | No | ||
| 64 | PITX2 | 7892 | 0.703 | 0.0349 | No | ||
| 65 | PDZRN4 | 7902 | 0.702 | 0.0357 | No | ||
| 66 | TCL1A | 7980 | 0.671 | 0.0333 | No | ||
| 67 | POU2F1 | 7999 | 0.663 | 0.0336 | No | ||
| 68 | PRKAB1 | 8148 | 0.596 | 0.0279 | No | ||
| 69 | LMO3 | 8247 | 0.558 | 0.0243 | No | ||
| 70 | TITF1 | 8418 | 0.491 | 0.0174 | No | ||
| 71 | NRL | 8537 | 0.439 | 0.0127 | No | ||
| 72 | GNG8 | 8718 | 0.356 | 0.0051 | No | ||
| 73 | NAP1L5 | 8774 | 0.331 | 0.0031 | No | ||
| 74 | GPRC5D | 9323 | 0.097 | -0.0219 | No | ||
| 75 | CRISP1 | 9544 | -0.013 | -0.0320 | No | ||
| 76 | WNT10A | 9640 | -0.061 | -0.0363 | No | ||
| 77 | CXXC6 | 9726 | -0.096 | -0.0400 | No | ||
| 78 | HOXB6 | 9880 | -0.167 | -0.0467 | No | ||
| 79 | DLX5 | 9964 | -0.203 | -0.0502 | No | ||
| 80 | PNOC | 10333 | -0.352 | -0.0665 | No | ||
| 81 | TBR1 | 10724 | -0.484 | -0.0836 | No | ||
| 82 | KCTD12 | 11147 | -0.636 | -0.1019 | No | ||
| 83 | ARRDC3 | 11277 | -0.676 | -0.1066 | No | ||
| 84 | HOXC4 | 11324 | -0.689 | -0.1075 | No | ||
| 85 | ATOH1 | 11388 | -0.710 | -0.1092 | No | ||
| 86 | EREG | 11573 | -0.777 | -0.1163 | No | ||
| 87 | ASB7 | 11688 | -0.820 | -0.1201 | No | ||
| 88 | LRP1B | 11748 | -0.841 | -0.1213 | No | ||
| 89 | SLC6A15 | 12049 | -0.935 | -0.1335 | No | ||
| 90 | ESRRG | 12112 | -0.951 | -0.1347 | No | ||
| 91 | KCNH5 | 12154 | -0.962 | -0.1349 | No | ||
| 92 | TBX19 | 12303 | -1.007 | -0.1400 | No | ||
| 93 | DOCK4 | 12374 | -1.030 | -0.1414 | No | ||
| 94 | ZDHHC21 | 12495 | -1.067 | -0.1450 | No | ||
| 95 | TBXAS1 | 12507 | -1.069 | -0.1437 | No | ||
| 96 | SEZ6 | 12840 | -1.168 | -0.1569 | No | ||
| 97 | ANLN | 13041 | -1.230 | -0.1639 | No | ||
| 98 | GAD1 | 13194 | -1.272 | -0.1687 | No | ||
| 99 | GPC4 | 13223 | -1.281 | -0.1678 | No | ||
| 100 | WNT8B | 13322 | -1.308 | -0.1700 | No | ||
| 101 | EN1 | 13659 | -1.411 | -0.1830 | No | ||
| 102 | SLC6A5 | 13826 | -1.458 | -0.1881 | No | ||
| 103 | CX36 | 14323 | -1.603 | -0.2081 | No | ||
| 104 | HSD17B2 | 14410 | -1.623 | -0.2092 | No | ||
| 105 | SULF1 | 14569 | -1.671 | -0.2135 | No | ||
| 106 | HMGA2 | 14593 | -1.677 | -0.2117 | No | ||
| 107 | HOXC5 | 14622 | -1.689 | -0.2100 | No | ||
| 108 | KCTD15 | 14702 | -1.713 | -0.2107 | No | ||
| 109 | HTR3B | 14762 | -1.730 | -0.2103 | No | ||
| 110 | CER1 | 14866 | -1.760 | -0.2120 | No | ||
| 111 | DNAJC5B | 14884 | -1.765 | -0.2097 | No | ||
| 112 | CA4 | 14915 | -1.775 | -0.2080 | No | ||
| 113 | NELL2 | 15270 | -1.884 | -0.2210 | No | ||
| 114 | GJA1 | 15279 | -1.888 | -0.2181 | No | ||
| 115 | UBQLN1 | 15467 | -1.946 | -0.2233 | No | ||
| 116 | FGF7 | 15563 | -1.987 | -0.2242 | No | ||
| 117 | HOXC6 | 15663 | -2.021 | -0.2252 | No | ||
| 118 | PPP1R14C | 15688 | -2.030 | -0.2228 | No | ||
| 119 | HSPB3 | 15694 | -2.030 | -0.2194 | No | ||
| 120 | LRRC15 | 15853 | -2.080 | -0.2231 | No | ||
| 121 | COL12A1 | 15878 | -2.087 | -0.2205 | No | ||
| 122 | RPL10 | 15899 | -2.094 | -0.2178 | No | ||
| 123 | PHEX | 16375 | -2.241 | -0.2357 | No | ||
| 124 | FGF10 | 16511 | -2.292 | -0.2379 | No | ||
| 125 | HAS2 | 16601 | -2.323 | -0.2380 | No | ||
| 126 | DMD | 16796 | -2.392 | -0.2427 | No | ||
| 127 | UBE2E2 | 16927 | -2.442 | -0.2444 | No | ||
| 128 | CXCL14 | 16943 | -2.447 | -0.2409 | No | ||
| 129 | FLRT2 | 17076 | -2.491 | -0.2426 | No | ||
| 130 | NEUROG1 | 17222 | -2.543 | -0.2448 | Yes | ||
| 131 | DPF3 | 17243 | -2.551 | -0.2413 | Yes | ||
| 132 | PROK1 | 17274 | -2.564 | -0.2382 | Yes | ||
| 133 | HOXD8 | 17337 | -2.592 | -0.2365 | Yes | ||
| 134 | ELAVL2 | 17342 | -2.594 | -0.2322 | Yes | ||
| 135 | SHOX2 | 17350 | -2.597 | -0.2280 | Yes | ||
| 136 | SLC4A4 | 17552 | -2.664 | -0.2326 | Yes | ||
| 137 | LTBP1 | 17640 | -2.702 | -0.2319 | Yes | ||
| 138 | HHIP | 17668 | -2.713 | -0.2284 | Yes | ||
| 139 | ANK3 | 17794 | -2.770 | -0.2293 | Yes | ||
| 140 | RWDD2 | 17837 | -2.787 | -0.2264 | Yes | ||
| 141 | HESX1 | 17852 | -2.792 | -0.2221 | Yes | ||
| 142 | FGF3 | 17884 | -2.804 | -0.2187 | Yes | ||
| 143 | FOXP2 | 17925 | -2.821 | -0.2156 | Yes | ||
| 144 | RORA | 17932 | -2.824 | -0.2109 | Yes | ||
| 145 | NHLH2 | 18159 | -2.920 | -0.2162 | Yes | ||
| 146 | PVRL3 | 18175 | -2.927 | -0.2118 | Yes | ||
| 147 | HDAC9 | 18201 | -2.936 | -0.2079 | Yes | ||
| 148 | PRRX1 | 18256 | -2.957 | -0.2052 | Yes | ||
| 149 | PCDH9 | 18304 | -2.978 | -0.2021 | Yes | ||
| 150 | ACP6 | 18461 | -3.054 | -0.2040 | Yes | ||
| 151 | SLC16A6 | 18482 | -3.064 | -0.1996 | Yes | ||
| 152 | MBNL2 | 18541 | -3.091 | -0.1968 | Yes | ||
| 153 | NTRK2 | 18614 | -3.135 | -0.1947 | Yes | ||
| 154 | PAX3 | 18724 | -3.198 | -0.1941 | Yes | ||
| 155 | INA | 18943 | -3.323 | -0.1983 | Yes | ||
| 156 | MINA | 19014 | -3.361 | -0.1957 | Yes | ||
| 157 | CHCHD7 | 19061 | -3.388 | -0.1919 | Yes | ||
| 158 | GPR65 | 19252 | -3.510 | -0.1945 | Yes | ||
| 159 | PPFIBP1 | 19256 | -3.512 | -0.1885 | Yes | ||
| 160 | RALGPS2 | 19561 | -3.736 | -0.1960 | Yes | ||
| 161 | RNF11 | 19674 | -3.827 | -0.1945 | Yes | ||
| 162 | IMPDH2 | 19737 | -3.887 | -0.1905 | Yes | ||
| 163 | CART1 | 19761 | -3.907 | -0.1848 | Yes | ||
| 164 | VGLL1 | 19763 | -3.910 | -0.1780 | Yes | ||
| 165 | DLX1 | 19783 | -3.932 | -0.1720 | Yes | ||
| 166 | INPPL1 | 19844 | -3.988 | -0.1678 | Yes | ||
| 167 | HTR2C | 19897 | -4.029 | -0.1632 | Yes | ||
| 168 | PSCD3 | 20086 | -4.223 | -0.1645 | Yes | ||
| 169 | SLC16A14 | 20108 | -4.252 | -0.1580 | Yes | ||
| 170 | EYA1 | 20145 | -4.286 | -0.1522 | Yes | ||
| 171 | TPM2 | 20185 | -4.335 | -0.1464 | Yes | ||
| 172 | SEZ6L | 20304 | -4.450 | -0.1441 | Yes | ||
| 173 | NEO1 | 20318 | -4.470 | -0.1369 | Yes | ||
| 174 | KLHL13 | 20504 | -4.728 | -0.1372 | Yes | ||
| 175 | CACNA1C | 20533 | -4.759 | -0.1301 | Yes | ||
| 176 | LRP1 | 20637 | -4.891 | -0.1263 | Yes | ||
| 177 | KLHL1 | 20638 | -4.894 | -0.1178 | Yes | ||
| 178 | PTPN21 | 20727 | -5.076 | -0.1130 | Yes | ||
| 179 | PPAP2B | 20759 | -5.127 | -0.1055 | Yes | ||
| 180 | LMO4 | 20804 | -5.199 | -0.0984 | Yes | ||
| 181 | MAB21L2 | 20881 | -5.298 | -0.0927 | Yes | ||
| 182 | UBXD3 | 20939 | -5.375 | -0.0859 | Yes | ||
| 183 | DAB1 | 20954 | -5.399 | -0.0772 | Yes | ||
| 184 | CXXC4 | 21019 | -5.527 | -0.0705 | Yes | ||
| 185 | HOXA3 | 21069 | -5.643 | -0.0629 | Yes | ||
| 186 | SFRS7 | 21128 | -5.770 | -0.0555 | Yes | ||
| 187 | CITED2 | 21215 | -6.023 | -0.0489 | Yes | ||
| 188 | GNA13 | 21224 | -6.051 | -0.0388 | Yes | ||
| 189 | TSGA14 | 21279 | -6.214 | -0.0304 | Yes | ||
| 190 | JDP2 | 21351 | -6.434 | -0.0224 | Yes | ||
| 191 | DNMT3B | 21694 | -8.012 | -0.0242 | Yes | ||
| 192 | NOTCH1 | 21772 | -8.813 | -0.0124 | Yes | ||
| 193 | TRPM1 | 21891 | -11.674 | 0.0026 | Yes |
