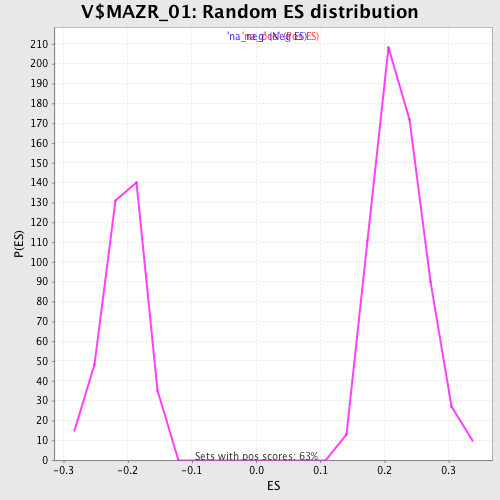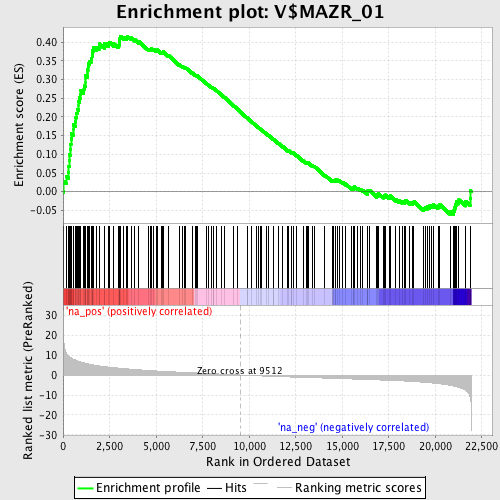
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$MAZR_01 |
| Enrichment Score (ES) | 0.4160804 |
| Normalized Enrichment Score (NES) | 1.8630522 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.009235111 |
| FWER p-Value | 0.066 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | SSBP2 | 36 | 16.873 | 0.0281 | Yes | ||
| 2 | UTX | 181 | 10.828 | 0.0406 | Yes | ||
| 3 | BAZ2A | 293 | 9.571 | 0.0523 | Yes | ||
| 4 | FOXJ2 | 307 | 9.460 | 0.0684 | Yes | ||
| 5 | THRAP1 | 337 | 9.200 | 0.0833 | Yes | ||
| 6 | PHF12 | 345 | 9.131 | 0.0991 | Yes | ||
| 7 | JUNB | 376 | 8.916 | 0.1134 | Yes | ||
| 8 | TNPO2 | 406 | 8.708 | 0.1274 | Yes | ||
| 9 | KCNH2 | 427 | 8.532 | 0.1415 | Yes | ||
| 10 | RFX1 | 446 | 8.462 | 0.1556 | Yes | ||
| 11 | RTN3 | 542 | 7.957 | 0.1653 | Yes | ||
| 12 | TCF7L2 | 546 | 7.944 | 0.1792 | Yes | ||
| 13 | SLC4A2 | 665 | 7.427 | 0.1868 | Yes | ||
| 14 | SRF | 686 | 7.344 | 0.1989 | Yes | ||
| 15 | PAFAH1B1 | 726 | 7.153 | 0.2097 | Yes | ||
| 16 | RAB1B | 751 | 7.053 | 0.2210 | Yes | ||
| 17 | RAB11B | 821 | 6.811 | 0.2298 | Yes | ||
| 18 | TRIM41 | 824 | 6.804 | 0.2417 | Yes | ||
| 19 | POGZ | 876 | 6.646 | 0.2511 | Yes | ||
| 20 | SF1 | 922 | 6.531 | 0.2606 | Yes | ||
| 21 | FBS1 | 937 | 6.504 | 0.2714 | Yes | ||
| 22 | GTPBP2 | 1119 | 6.047 | 0.2737 | Yes | ||
| 23 | DNMT3A | 1150 | 5.973 | 0.2829 | Yes | ||
| 24 | VASP | 1190 | 5.873 | 0.2914 | Yes | ||
| 25 | MKNK2 | 1193 | 5.865 | 0.3017 | Yes | ||
| 26 | UBE1 | 1195 | 5.864 | 0.3120 | Yes | ||
| 27 | DDAH2 | 1319 | 5.569 | 0.3161 | Yes | ||
| 28 | ENSA | 1323 | 5.567 | 0.3258 | Yes | ||
| 29 | MBD6 | 1339 | 5.529 | 0.3349 | Yes | ||
| 30 | UBE2R2 | 1356 | 5.488 | 0.3438 | Yes | ||
| 31 | MLL5 | 1438 | 5.327 | 0.3495 | Yes | ||
| 32 | RGL2 | 1501 | 5.211 | 0.3558 | Yes | ||
| 33 | GABARAPL2 | 1552 | 5.124 | 0.3626 | Yes | ||
| 34 | CLIC1 | 1566 | 5.097 | 0.3709 | Yes | ||
| 35 | TLK2 | 1596 | 5.060 | 0.3785 | Yes | ||
| 36 | NOTCH2 | 1650 | 4.966 | 0.3848 | Yes | ||
| 37 | CHD4 | 1814 | 4.734 | 0.3857 | Yes | ||
| 38 | UBE4B | 1934 | 4.560 | 0.3883 | Yes | ||
| 39 | RKHD3 | 1958 | 4.524 | 0.3952 | Yes | ||
| 40 | MTHFR | 2216 | 4.207 | 0.3908 | Yes | ||
| 41 | FUS | 2229 | 4.200 | 0.3977 | Yes | ||
| 42 | NLK | 2430 | 3.973 | 0.3955 | Yes | ||
| 43 | ROCK1 | 2484 | 3.908 | 0.3999 | Yes | ||
| 44 | LUC7L2 | 2721 | 3.683 | 0.3956 | Yes | ||
| 45 | DMPK | 2956 | 3.470 | 0.3910 | Yes | ||
| 46 | ITGA6 | 3003 | 3.432 | 0.3949 | Yes | ||
| 47 | TRPS1 | 3024 | 3.417 | 0.4000 | Yes | ||
| 48 | PCF11 | 3029 | 3.410 | 0.4058 | Yes | ||
| 49 | HNRPR | 3052 | 3.395 | 0.4108 | Yes | ||
| 50 | ATF2 | 3068 | 3.373 | 0.4161 | Yes | ||
| 51 | TRERF1 | 3248 | 3.224 | 0.4135 | No | ||
| 52 | CBFB | 3407 | 3.097 | 0.4118 | No | ||
| 53 | RAB5C | 3461 | 3.051 | 0.4147 | No | ||
| 54 | NFYC | 3647 | 2.909 | 0.4113 | No | ||
| 55 | CDC27 | 3832 | 2.779 | 0.4078 | No | ||
| 56 | GNB2 | 4074 | 2.605 | 0.4013 | No | ||
| 57 | PTPRF | 4606 | 2.258 | 0.3809 | No | ||
| 58 | EZH2 | 4701 | 2.198 | 0.3805 | No | ||
| 59 | ABR | 4741 | 2.179 | 0.3825 | No | ||
| 60 | RNF111 | 4856 | 2.118 | 0.3810 | No | ||
| 61 | HSD11B1 | 4993 | 2.042 | 0.3784 | No | ||
| 62 | SEPT3 | 5053 | 2.001 | 0.3792 | No | ||
| 63 | NCOA2 | 5268 | 1.893 | 0.3727 | No | ||
| 64 | UST | 5324 | 1.865 | 0.3735 | No | ||
| 65 | SP1 | 5374 | 1.845 | 0.3745 | No | ||
| 66 | BAT2 | 5654 | 1.700 | 0.3647 | No | ||
| 67 | IL1RAPL1 | 6231 | 1.436 | 0.3408 | No | ||
| 68 | HNRPUL1 | 6409 | 1.356 | 0.3350 | No | ||
| 69 | CLCN6 | 6505 | 1.306 | 0.3330 | No | ||
| 70 | FKBP2 | 6598 | 1.258 | 0.3310 | No | ||
| 71 | NTN2L | 6931 | 1.109 | 0.3177 | No | ||
| 72 | NLGN2 | 7107 | 1.035 | 0.3115 | No | ||
| 73 | ZBTB26 | 7155 | 1.017 | 0.3111 | No | ||
| 74 | TRIP10 | 7208 | 0.995 | 0.3105 | No | ||
| 75 | FXYD1 | 7678 | 0.793 | 0.2903 | No | ||
| 76 | ENTPD1 | 7818 | 0.731 | 0.2852 | No | ||
| 77 | NETO1 | 7977 | 0.672 | 0.2792 | No | ||
| 78 | PIM1 | 8068 | 0.635 | 0.2762 | No | ||
| 79 | SON | 8100 | 0.621 | 0.2758 | No | ||
| 80 | DLL3 | 8220 | 0.569 | 0.2714 | No | ||
| 81 | NXPH4 | 8515 | 0.446 | 0.2587 | No | ||
| 82 | FBXL19 | 8665 | 0.384 | 0.2525 | No | ||
| 83 | LGALS1 | 9176 | 0.161 | 0.2294 | No | ||
| 84 | SLC39A5 | 9368 | 0.073 | 0.2207 | No | ||
| 85 | HOXB6 | 9880 | -0.167 | 0.1976 | No | ||
| 86 | JPH4 | 9886 | -0.168 | 0.1976 | No | ||
| 87 | PLA2G2E | 10106 | -0.264 | 0.1881 | No | ||
| 88 | GRIN2D | 10400 | -0.373 | 0.1753 | No | ||
| 89 | SULT2B1 | 10506 | -0.406 | 0.1712 | No | ||
| 90 | FXYD3 | 10599 | -0.439 | 0.1677 | No | ||
| 91 | MYBPH | 10636 | -0.451 | 0.1668 | No | ||
| 92 | CTNNBIP1 | 10901 | -0.548 | 0.1557 | No | ||
| 93 | MBD3 | 11026 | -0.588 | 0.1510 | No | ||
| 94 | HMX1 | 11296 | -0.680 | 0.1399 | No | ||
| 95 | HOXC4 | 11324 | -0.689 | 0.1399 | No | ||
| 96 | HOXB9 | 11583 | -0.779 | 0.1294 | No | ||
| 97 | GATA6 | 11803 | -0.857 | 0.1209 | No | ||
| 98 | SALL1 | 12077 | -0.942 | 0.1100 | No | ||
| 99 | RAP2C | 12125 | -0.957 | 0.1095 | No | ||
| 100 | BAI2 | 12283 | -1.002 | 0.1041 | No | ||
| 101 | NPTX1 | 12293 | -1.004 | 0.1054 | No | ||
| 102 | SLITRK5 | 12368 | -1.029 | 0.1038 | No | ||
| 103 | LRRTM4 | 12556 | -1.084 | 0.0972 | No | ||
| 104 | MOV10 | 12911 | -1.190 | 0.0830 | No | ||
| 105 | DUSP8 | 13082 | -1.240 | 0.0774 | No | ||
| 106 | AEBP2 | 13141 | -1.258 | 0.0770 | No | ||
| 107 | KCND3 | 13189 | -1.271 | 0.0770 | No | ||
| 108 | LIN28 | 13412 | -1.335 | 0.0692 | No | ||
| 109 | TCF1 | 13483 | -1.356 | 0.0684 | No | ||
| 110 | EIF4G1 | 14061 | -1.526 | 0.0446 | No | ||
| 111 | NEUROD2 | 14457 | -1.636 | 0.0293 | No | ||
| 112 | LRFN5 | 14484 | -1.641 | 0.0310 | No | ||
| 113 | DLL4 | 14548 | -1.664 | 0.0311 | No | ||
| 114 | GGN | 14639 | -1.693 | 0.0299 | No | ||
| 115 | SLC39A2 | 14660 | -1.700 | 0.0320 | No | ||
| 116 | HR | 14740 | -1.724 | 0.0314 | No | ||
| 117 | ZNF462 | 14831 | -1.751 | 0.0304 | No | ||
| 118 | FGF17 | 15033 | -1.809 | 0.0243 | No | ||
| 119 | RTN2 | 15178 | -1.860 | 0.0210 | No | ||
| 120 | SYT3 | 15512 | -1.964 | 0.0092 | No | ||
| 121 | CSPG2 | 15606 | -2.002 | 0.0084 | No | ||
| 122 | CACNB1 | 15623 | -2.006 | 0.0112 | No | ||
| 123 | HOXC6 | 15663 | -2.021 | 0.0130 | No | ||
| 124 | THPO | 15822 | -2.070 | 0.0094 | No | ||
| 125 | MAGED2 | 15954 | -2.109 | 0.0071 | No | ||
| 126 | AHR | 16096 | -2.150 | 0.0044 | No | ||
| 127 | HPCA | 16348 | -2.233 | -0.0032 | No | ||
| 128 | TCEB1 | 16349 | -2.234 | 0.0008 | No | ||
| 129 | ATF1 | 16376 | -2.241 | 0.0035 | No | ||
| 130 | PAX6 | 16454 | -2.272 | 0.0040 | No | ||
| 131 | POU3F2 | 16862 | -2.419 | -0.0104 | No | ||
| 132 | SLC22A17 | 16905 | -2.432 | -0.0081 | No | ||
| 133 | APBA1 | 16935 | -2.444 | -0.0051 | No | ||
| 134 | FGF14 | 17196 | -2.532 | -0.0126 | No | ||
| 135 | CLOCK | 17259 | -2.556 | -0.0109 | No | ||
| 136 | STC2 | 17311 | -2.578 | -0.0087 | No | ||
| 137 | GRK5 | 17526 | -2.656 | -0.0138 | No | ||
| 138 | SLC2A4 | 17567 | -2.667 | -0.0110 | No | ||
| 139 | ZIC4 | 17876 | -2.800 | -0.0202 | No | ||
| 140 | PACSIN3 | 18067 | -2.885 | -0.0238 | No | ||
| 141 | HMGA1 | 18236 | -2.950 | -0.0263 | No | ||
| 142 | EFNA1 | 18342 | -2.998 | -0.0259 | No | ||
| 143 | MAPT | 18384 | -3.015 | -0.0224 | No | ||
| 144 | PARD6G | 18619 | -3.136 | -0.0276 | No | ||
| 145 | FOXA1 | 18781 | -3.226 | -0.0293 | No | ||
| 146 | EIF5A | 18853 | -3.265 | -0.0269 | No | ||
| 147 | SEMA4C | 19380 | -3.598 | -0.0447 | No | ||
| 148 | WNT1 | 19449 | -3.646 | -0.0414 | No | ||
| 149 | CORO1C | 19560 | -3.735 | -0.0398 | No | ||
| 150 | GRIK1 | 19670 | -3.825 | -0.0381 | No | ||
| 151 | H1F0 | 19790 | -3.935 | -0.0366 | No | ||
| 152 | KCNK6 | 19915 | -4.048 | -0.0352 | No | ||
| 153 | EYA1 | 20145 | -4.286 | -0.0381 | No | ||
| 154 | LRRN6A | 20245 | -4.397 | -0.0349 | No | ||
| 155 | HNRPDL | 20813 | -5.210 | -0.0518 | No | ||
| 156 | MTF1 | 20995 | -5.475 | -0.0504 | No | ||
| 157 | ETF1 | 21034 | -5.559 | -0.0424 | No | ||
| 158 | RBL1 | 21082 | -5.671 | -0.0345 | No | ||
| 159 | ETV5 | 21115 | -5.739 | -0.0259 | No | ||
| 160 | CAMK2G | 21269 | -6.196 | -0.0220 | No | ||
| 161 | EPHB2 | 21625 | -7.511 | -0.0250 | No | ||
| 162 | GART | 21869 | -10.928 | -0.0169 | No | ||
| 163 | PCBP4 | 21889 | -11.615 | 0.0027 | No |