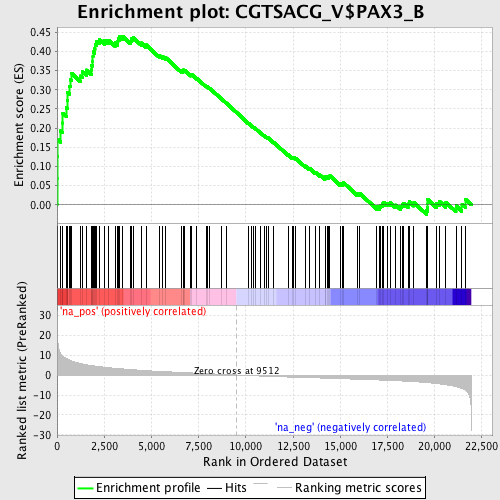
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CGTSACG_V$PAX3_B |
| Enrichment Score (ES) | 0.43976307 |
| Normalized Enrichment Score (NES) | 1.8265817 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0086735925 |
| FWER p-Value | 0.104 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EGR3 | 3 | 25.122 | 0.0682 | Yes | ||
| 2 | XBP1 | 6 | 21.674 | 0.1270 | Yes | ||
| 3 | EGR2 | 41 | 16.374 | 0.1700 | Yes | ||
| 4 | AP2B1 | 186 | 10.759 | 0.1926 | Yes | ||
| 5 | CRY1 | 301 | 9.519 | 0.2133 | Yes | ||
| 6 | RAD9A | 304 | 9.493 | 0.2390 | Yes | ||
| 7 | BCL11B | 482 | 8.269 | 0.2534 | Yes | ||
| 8 | PIAS4 | 537 | 7.975 | 0.2726 | Yes | ||
| 9 | EGR1 | 548 | 7.925 | 0.2937 | Yes | ||
| 10 | WDR23 | 670 | 7.418 | 0.3084 | Yes | ||
| 11 | NAB2 | 695 | 7.297 | 0.3271 | Yes | ||
| 12 | GNAS | 780 | 6.945 | 0.3421 | Yes | ||
| 13 | ZZEF1 | 1258 | 5.727 | 0.3359 | Yes | ||
| 14 | PNRC1 | 1331 | 5.552 | 0.3477 | Yes | ||
| 15 | ANKHD1 | 1553 | 5.123 | 0.3515 | Yes | ||
| 16 | RELB | 1827 | 4.711 | 0.3518 | Yes | ||
| 17 | HTATIP | 1838 | 4.694 | 0.3641 | Yes | ||
| 18 | CBX6 | 1891 | 4.608 | 0.3743 | Yes | ||
| 19 | POLB | 1899 | 4.594 | 0.3864 | Yes | ||
| 20 | PPP2R1A | 1905 | 4.589 | 0.3987 | Yes | ||
| 21 | PB1 | 2000 | 4.460 | 0.4065 | Yes | ||
| 22 | NDUFB2 | 2008 | 4.451 | 0.4183 | Yes | ||
| 23 | EIF4E | 2088 | 4.365 | 0.4265 | Yes | ||
| 24 | NR4A1 | 2240 | 4.181 | 0.4310 | Yes | ||
| 25 | SLC38A2 | 2520 | 3.874 | 0.4288 | Yes | ||
| 26 | EPC1 | 2723 | 3.682 | 0.4295 | Yes | ||
| 27 | BCL11A | 3071 | 3.369 | 0.4228 | Yes | ||
| 28 | PPT2 | 3219 | 3.247 | 0.4249 | Yes | ||
| 29 | SOAT1 | 3227 | 3.240 | 0.4334 | Yes | ||
| 30 | PTPN23 | 3320 | 3.165 | 0.4378 | Yes | ||
| 31 | BRAF | 3459 | 3.052 | 0.4398 | Yes | ||
| 32 | NR4A2 | 3907 | 2.716 | 0.4267 | No | ||
| 33 | EDEM1 | 3938 | 2.692 | 0.4326 | No | ||
| 34 | CTCF | 4032 | 2.627 | 0.4355 | No | ||
| 35 | CUL5 | 4468 | 2.343 | 0.4220 | No | ||
| 36 | TNRC15 | 4710 | 2.195 | 0.4169 | No | ||
| 37 | RAD51C | 5406 | 1.830 | 0.3901 | No | ||
| 38 | HMOX2 | 5579 | 1.736 | 0.3869 | No | ||
| 39 | HPS5 | 5756 | 1.659 | 0.3834 | No | ||
| 40 | USP8 | 6578 | 1.271 | 0.3493 | No | ||
| 41 | HOXC10 | 6671 | 1.224 | 0.3484 | No | ||
| 42 | GEM | 6673 | 1.223 | 0.3517 | No | ||
| 43 | SMARCA5 | 6755 | 1.192 | 0.3512 | No | ||
| 44 | YME1L1 | 7093 | 1.040 | 0.3386 | No | ||
| 45 | SLC18A2 | 7129 | 1.026 | 0.3398 | No | ||
| 46 | NRK | 7397 | 0.910 | 0.3300 | No | ||
| 47 | PITX2 | 7892 | 0.703 | 0.3093 | No | ||
| 48 | PPP1R3D | 7966 | 0.675 | 0.3078 | No | ||
| 49 | TEX14 | 8081 | 0.628 | 0.3043 | No | ||
| 50 | WSB1 | 8723 | 0.353 | 0.2759 | No | ||
| 51 | GTF2H1 | 8990 | 0.237 | 0.2644 | No | ||
| 52 | EN2 | 10148 | -0.278 | 0.2122 | No | ||
| 53 | PAK2 | 10297 | -0.335 | 0.2063 | No | ||
| 54 | KCNE4 | 10405 | -0.375 | 0.2024 | No | ||
| 55 | IRX6 | 10512 | -0.408 | 0.1987 | No | ||
| 56 | SLC6A12 | 10771 | -0.503 | 0.1883 | No | ||
| 57 | RNF146 | 10969 | -0.572 | 0.1808 | No | ||
| 58 | TSPYL5 | 11111 | -0.623 | 0.1760 | No | ||
| 59 | EGR4 | 11183 | -0.648 | 0.1746 | No | ||
| 60 | PPM2C | 11477 | -0.742 | 0.1632 | No | ||
| 61 | RBBP8 | 12250 | -0.991 | 0.1305 | No | ||
| 62 | FOXD3 | 12457 | -1.052 | 0.1239 | No | ||
| 63 | BHLHB2 | 12513 | -1.071 | 0.1243 | No | ||
| 64 | STXBP1 | 12638 | -1.106 | 0.1217 | No | ||
| 65 | PEPD | 13159 | -1.262 | 0.1013 | No | ||
| 66 | ADAM10 | 13366 | -1.321 | 0.0955 | No | ||
| 67 | RASD1 | 13675 | -1.418 | 0.0852 | No | ||
| 68 | NOL4 | 13922 | -1.484 | 0.0780 | No | ||
| 69 | ZZZ3 | 14196 | -1.566 | 0.0698 | No | ||
| 70 | NKX2-3 | 14220 | -1.573 | 0.0730 | No | ||
| 71 | CX36 | 14323 | -1.603 | 0.0727 | No | ||
| 72 | PHACTR3 | 14403 | -1.621 | 0.0735 | No | ||
| 73 | NEUROD2 | 14457 | -1.636 | 0.0755 | No | ||
| 74 | CDC14B | 14992 | -1.799 | 0.0559 | No | ||
| 75 | DHX36 | 15134 | -1.847 | 0.0545 | No | ||
| 76 | HOXA9 | 15182 | -1.861 | 0.0574 | No | ||
| 77 | CHGB | 15923 | -2.101 | 0.0292 | No | ||
| 78 | SULT4A1 | 16038 | -2.133 | 0.0298 | No | ||
| 79 | MAB21L1 | 16941 | -2.445 | -0.0048 | No | ||
| 80 | TRIB1 | 17100 | -2.499 | -0.0052 | No | ||
| 81 | GGA1 | 17159 | -2.518 | -0.0011 | No | ||
| 82 | PCNP | 17236 | -2.549 | 0.0024 | No | ||
| 83 | IPO7 | 17313 | -2.578 | 0.0059 | No | ||
| 84 | PLK2 | 17504 | -2.650 | 0.0044 | No | ||
| 85 | LTBP1 | 17640 | -2.702 | 0.0056 | No | ||
| 86 | HMGN1 | 17940 | -2.829 | -0.0004 | No | ||
| 87 | SUPT16H | 18208 | -2.939 | -0.0046 | No | ||
| 88 | CDKN1A | 18276 | -2.964 | 0.0004 | No | ||
| 89 | PTMA | 18360 | -3.006 | 0.0047 | No | ||
| 90 | ZFHX1B | 18599 | -3.124 | 0.0023 | No | ||
| 91 | CANX | 18647 | -3.149 | 0.0088 | No | ||
| 92 | ELOVL5 | 18907 | -3.302 | 0.0059 | No | ||
| 93 | IKBKB | 19586 | -3.755 | -0.0149 | No | ||
| 94 | RAB7 | 19628 | -3.788 | -0.0065 | No | ||
| 95 | CASK | 19642 | -3.801 | 0.0032 | No | ||
| 96 | RBM18 | 19644 | -3.801 | 0.0135 | No | ||
| 97 | SMARCE1 | 20090 | -4.224 | 0.0046 | No | ||
| 98 | MASTL | 20242 | -4.396 | 0.0097 | No | ||
| 99 | EHD4 | 20601 | -4.840 | 0.0064 | No | ||
| 100 | MRRF | 21138 | -5.800 | -0.0023 | No | ||
| 101 | TAF11 | 21454 | -6.783 | 0.0017 | No | ||
| 102 | SLC25A3 | 21648 | -7.658 | 0.0137 | No |
