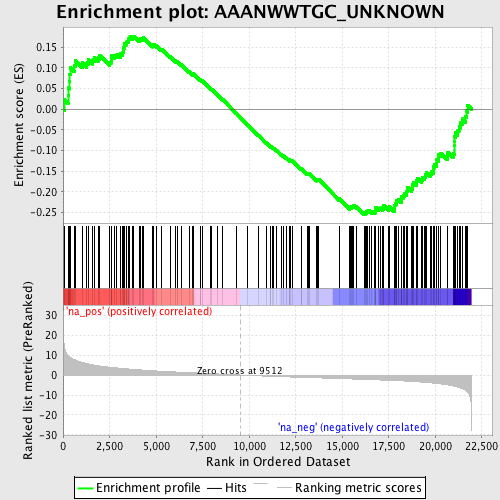
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | AAANWWTGC_UNKNOWN |
| Enrichment Score (ES) | -0.25492886 |
| Normalized Enrichment Score (NES) | -1.2320204 |
| Nominal p-value | 0.047058824 |
| FDR q-value | 0.4388611 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BCL6 | 77 | 13.547 | 0.0232 | No | ||
| 2 | RBMX | 267 | 9.771 | 0.0337 | No | ||
| 3 | NTN1 | 292 | 9.572 | 0.0515 | No | ||
| 4 | DUSP1 | 351 | 9.094 | 0.0668 | No | ||
| 5 | ACTB | 360 | 9.014 | 0.0841 | No | ||
| 6 | CHES1 | 390 | 8.863 | 0.1003 | No | ||
| 7 | PPP1R10 | 600 | 7.677 | 0.1058 | No | ||
| 8 | WDR23 | 670 | 7.418 | 0.1173 | No | ||
| 9 | FOXP1 | 1045 | 6.223 | 0.1124 | No | ||
| 10 | TLK1 | 1282 | 5.656 | 0.1127 | No | ||
| 11 | MAN2A2 | 1359 | 5.482 | 0.1200 | No | ||
| 12 | TLE4 | 1595 | 5.061 | 0.1192 | No | ||
| 13 | MYLK | 1683 | 4.921 | 0.1249 | No | ||
| 14 | CDC42EP3 | 1880 | 4.634 | 0.1250 | No | ||
| 15 | MAPK3 | 1966 | 4.511 | 0.1300 | No | ||
| 16 | SESN2 | 2486 | 3.907 | 0.1139 | No | ||
| 17 | KLF12 | 2575 | 3.833 | 0.1174 | No | ||
| 18 | CHD2 | 2595 | 3.816 | 0.1240 | No | ||
| 19 | MBNL1 | 2609 | 3.802 | 0.1309 | No | ||
| 20 | EIF4EBP2 | 2768 | 3.639 | 0.1309 | No | ||
| 21 | ATP1B1 | 2894 | 3.522 | 0.1321 | No | ||
| 22 | BCL11A | 3071 | 3.369 | 0.1306 | No | ||
| 23 | ZW10 | 3101 | 3.349 | 0.1359 | No | ||
| 24 | DNAJB5 | 3210 | 3.252 | 0.1373 | No | ||
| 25 | MYCL1 | 3237 | 3.234 | 0.1425 | No | ||
| 26 | SSBP3 | 3241 | 3.230 | 0.1487 | No | ||
| 27 | SORT1 | 3277 | 3.200 | 0.1534 | No | ||
| 28 | ANKRD15 | 3286 | 3.190 | 0.1594 | No | ||
| 29 | MAP3K4 | 3381 | 3.115 | 0.1612 | No | ||
| 30 | HOXB2 | 3430 | 3.081 | 0.1650 | No | ||
| 31 | SKIL | 3528 | 2.998 | 0.1665 | No | ||
| 32 | ELF4 | 3534 | 2.991 | 0.1722 | No | ||
| 33 | PPARGC1A | 3592 | 2.953 | 0.1754 | No | ||
| 34 | INHBA | 3744 | 2.844 | 0.1740 | No | ||
| 35 | RAB10 | 3803 | 2.800 | 0.1769 | No | ||
| 36 | ZHX3 | 4109 | 2.583 | 0.1680 | No | ||
| 37 | TRAM1 | 4146 | 2.550 | 0.1714 | No | ||
| 38 | MRPL24 | 4260 | 2.464 | 0.1710 | No | ||
| 39 | FBXW7 | 4301 | 2.440 | 0.1740 | No | ||
| 40 | DDIT3 | 4814 | 2.141 | 0.1547 | No | ||
| 41 | SOX4 | 4852 | 2.120 | 0.1572 | No | ||
| 42 | SNCAIP | 5007 | 2.028 | 0.1541 | No | ||
| 43 | GRIN2B | 5286 | 1.882 | 0.1451 | No | ||
| 44 | STEAP2 | 5746 | 1.665 | 0.1273 | No | ||
| 45 | FGFR2 | 6039 | 1.526 | 0.1169 | No | ||
| 46 | SKP2 | 6121 | 1.492 | 0.1161 | No | ||
| 47 | NRAS | 6347 | 1.384 | 0.1085 | No | ||
| 48 | DRD3 | 6791 | 1.177 | 0.0905 | No | ||
| 49 | PAX1 | 6972 | 1.094 | 0.0844 | No | ||
| 50 | SPAG9 | 6982 | 1.088 | 0.0861 | No | ||
| 51 | BNC2 | 7370 | 0.923 | 0.0702 | No | ||
| 52 | HHEX | 7379 | 0.918 | 0.0716 | No | ||
| 53 | SIX5 | 7462 | 0.883 | 0.0696 | No | ||
| 54 | OTX2 | 7936 | 0.690 | 0.0493 | No | ||
| 55 | POU2F1 | 7999 | 0.663 | 0.0477 | No | ||
| 56 | PCSK1 | 8309 | 0.532 | 0.0346 | No | ||
| 57 | CACNG3 | 8583 | 0.417 | 0.0229 | No | ||
| 58 | MYH2 | 9304 | 0.103 | -0.0099 | No | ||
| 59 | HOXB6 | 9880 | -0.167 | -0.0360 | No | ||
| 60 | GLRA2 | 10495 | -0.403 | -0.0634 | No | ||
| 61 | SALL3 | 10912 | -0.553 | -0.0814 | No | ||
| 62 | PPP2R2A | 11150 | -0.638 | -0.0910 | No | ||
| 63 | PHTF1 | 11154 | -0.640 | -0.0899 | No | ||
| 64 | SFRP2 | 11248 | -0.669 | -0.0928 | No | ||
| 65 | HOXC4 | 11324 | -0.689 | -0.0949 | No | ||
| 66 | CDC42EP5 | 11467 | -0.737 | -0.1000 | No | ||
| 67 | SPARCL1 | 11730 | -0.837 | -0.1104 | No | ||
| 68 | SGCD | 11827 | -0.866 | -0.1131 | No | ||
| 69 | SNX25 | 11977 | -0.910 | -0.1181 | No | ||
| 70 | SOX13 | 12143 | -0.960 | -0.1238 | No | ||
| 71 | SATB2 | 12145 | -0.960 | -0.1219 | No | ||
| 72 | PRIMA1 | 12216 | -0.981 | -0.1232 | No | ||
| 73 | KLF14 | 12304 | -1.008 | -0.1252 | No | ||
| 74 | LOX | 12782 | -1.153 | -0.1449 | No | ||
| 75 | ASPA | 12784 | -1.154 | -0.1426 | No | ||
| 76 | ANK2 | 13146 | -1.259 | -0.1567 | No | ||
| 77 | MID1 | 13180 | -1.269 | -0.1557 | No | ||
| 78 | GPC6 | 13233 | -1.283 | -0.1556 | No | ||
| 79 | ATP2B4 | 13634 | -1.402 | -0.1712 | No | ||
| 80 | KRTAP8-1 | 13654 | -1.408 | -0.1693 | No | ||
| 81 | PRDM16 | 13728 | -1.434 | -0.1698 | No | ||
| 82 | ZNF462 | 14831 | -1.751 | -0.2169 | No | ||
| 83 | DPYSL5 | 15400 | -1.925 | -0.2392 | No | ||
| 84 | PHOX2B | 15429 | -1.933 | -0.2367 | No | ||
| 85 | MGLL | 15496 | -1.960 | -0.2358 | No | ||
| 86 | FGF7 | 15563 | -1.987 | -0.2350 | No | ||
| 87 | SEMA6C | 15589 | -1.996 | -0.2322 | No | ||
| 88 | HOXA2 | 15771 | -2.054 | -0.2364 | No | ||
| 89 | ADHFE1 | 16175 | -2.173 | -0.2506 | Yes | ||
| 90 | ESR1 | 16252 | -2.203 | -0.2498 | Yes | ||
| 91 | NEK6 | 16305 | -2.219 | -0.2478 | Yes | ||
| 92 | MMP3 | 16371 | -2.239 | -0.2464 | Yes | ||
| 93 | PAX6 | 16454 | -2.272 | -0.2457 | Yes | ||
| 94 | PRKRIR | 16590 | -2.320 | -0.2473 | Yes | ||
| 95 | GANAB | 16718 | -2.362 | -0.2485 | Yes | ||
| 96 | EFNA5 | 16783 | -2.387 | -0.2467 | Yes | ||
| 97 | OMG | 16784 | -2.387 | -0.2420 | Yes | ||
| 98 | DMD | 16796 | -2.392 | -0.2378 | Yes | ||
| 99 | ZFPM1 | 16965 | -2.454 | -0.2407 | Yes | ||
| 100 | NNAT | 17039 | -2.479 | -0.2391 | Yes | ||
| 101 | NTRK3 | 17176 | -2.523 | -0.2404 | Yes | ||
| 102 | MEF2C | 17220 | -2.542 | -0.2374 | Yes | ||
| 103 | NEUROG1 | 17222 | -2.543 | -0.2324 | Yes | ||
| 104 | GPC3 | 17497 | -2.647 | -0.2398 | Yes | ||
| 105 | LIPG | 17540 | -2.659 | -0.2364 | Yes | ||
| 106 | ANK3 | 17794 | -2.770 | -0.2426 | Yes | ||
| 107 | ITM2C | 17808 | -2.776 | -0.2377 | Yes | ||
| 108 | EPHA7 | 17815 | -2.779 | -0.2325 | Yes | ||
| 109 | LSAMP | 17854 | -2.792 | -0.2288 | Yes | ||
| 110 | FOXP2 | 17925 | -2.821 | -0.2264 | Yes | ||
| 111 | RORA | 17932 | -2.824 | -0.2211 | Yes | ||
| 112 | PHF15 | 18005 | -2.856 | -0.2188 | Yes | ||
| 113 | PRPF4B | 18179 | -2.929 | -0.2210 | Yes | ||
| 114 | THBS2 | 18184 | -2.930 | -0.2154 | Yes | ||
| 115 | LOXL4 | 18198 | -2.935 | -0.2102 | Yes | ||
| 116 | TGIF | 18307 | -2.979 | -0.2093 | Yes | ||
| 117 | DSCAM | 18339 | -2.997 | -0.2048 | Yes | ||
| 118 | SHC3 | 18446 | -3.048 | -0.2037 | Yes | ||
| 119 | TLX3 | 18469 | -3.058 | -0.1987 | Yes | ||
| 120 | CDH13 | 18503 | -3.074 | -0.1941 | Yes | ||
| 121 | POU4F1 | 18516 | -3.081 | -0.1886 | Yes | ||
| 122 | PPFIA2 | 18742 | -3.208 | -0.1926 | Yes | ||
| 123 | CEPT1 | 18762 | -3.218 | -0.1871 | Yes | ||
| 124 | LRRN1 | 18785 | -3.228 | -0.1818 | Yes | ||
| 125 | RRS1 | 18837 | -3.257 | -0.1777 | Yes | ||
| 126 | CALM1 | 18993 | -3.349 | -0.1782 | Yes | ||
| 127 | CNTFR | 19002 | -3.356 | -0.1720 | Yes | ||
| 128 | FN1 | 19049 | -3.378 | -0.1674 | Yes | ||
| 129 | LECT1 | 19259 | -3.513 | -0.1701 | Yes | ||
| 130 | CD14 | 19303 | -3.540 | -0.1651 | Yes | ||
| 131 | SLMAP | 19442 | -3.642 | -0.1643 | Yes | ||
| 132 | SOX5 | 19470 | -3.663 | -0.1583 | Yes | ||
| 133 | PIK3R3 | 19528 | -3.708 | -0.1536 | Yes | ||
| 134 | PCTP | 19717 | -3.868 | -0.1546 | Yes | ||
| 135 | MEIS1 | 19781 | -3.929 | -0.1497 | Yes | ||
| 136 | GATA3 | 19901 | -4.038 | -0.1472 | Yes | ||
| 137 | DLGAP4 | 19907 | -4.040 | -0.1395 | Yes | ||
| 138 | ELAVL4 | 19973 | -4.101 | -0.1344 | Yes | ||
| 139 | DSTN | 20067 | -4.206 | -0.1304 | Yes | ||
| 140 | OLFM1 | 20081 | -4.221 | -0.1227 | Yes | ||
| 141 | FZD7 | 20160 | -4.299 | -0.1178 | Yes | ||
| 142 | TRPM3 | 20191 | -4.340 | -0.1106 | Yes | ||
| 143 | OLIG2 | 20284 | -4.431 | -0.1061 | Yes | ||
| 144 | NR2F2 | 20641 | -4.901 | -0.1128 | Yes | ||
| 145 | APP | 20670 | -4.957 | -0.1043 | Yes | ||
| 146 | DAB1 | 20954 | -5.399 | -0.1067 | Yes | ||
| 147 | PDGFRB | 21022 | -5.532 | -0.0988 | Yes | ||
| 148 | NFE2L2 | 21023 | -5.533 | -0.0879 | Yes | ||
| 149 | TFDP2 | 21031 | -5.552 | -0.0773 | Yes | ||
| 150 | GTF2E2 | 21039 | -5.579 | -0.0666 | Yes | ||
| 151 | HOXA3 | 21069 | -5.643 | -0.0569 | Yes | ||
| 152 | CITED2 | 21215 | -6.023 | -0.0516 | Yes | ||
| 153 | ATOH7 | 21274 | -6.201 | -0.0421 | Yes | ||
| 154 | MRPS18B | 21359 | -6.463 | -0.0332 | Yes | ||
| 155 | PPP3CC | 21439 | -6.721 | -0.0236 | Yes | ||
| 156 | EPHB2 | 21625 | -7.511 | -0.0173 | Yes | ||
| 157 | CDK2AP1 | 21670 | -7.801 | -0.0039 | Yes | ||
| 158 | CACNA1D | 21745 | -8.426 | 0.0093 | Yes |
