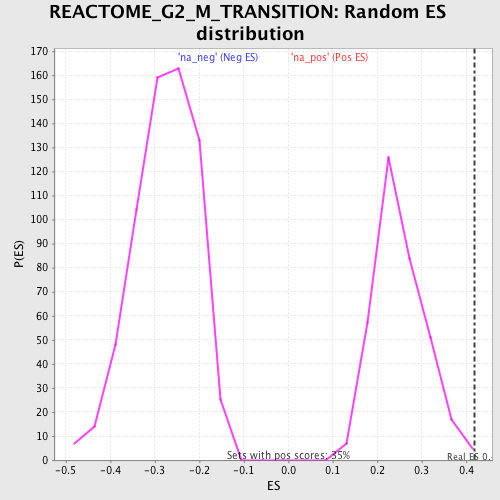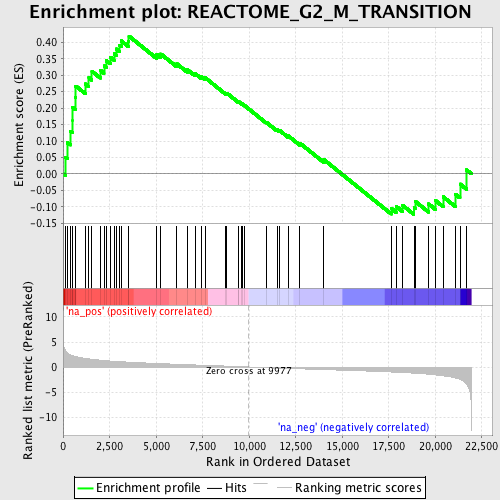
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_G2_M_TRANSITION |
| Enrichment Score (ES) | 0.41874105 |
| Normalized Enrichment Score (NES) | 1.6789124 |
| Nominal p-value | 0.0028818443 |
| FDR q-value | 0.31894445 |
| FWER p-Value | 0.986 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CCNH | 125 | 3.262 | 0.0507 | Yes | ||
| 2 | PCM1 | 229 | 2.821 | 0.0948 | Yes | ||
| 3 | NEK2 | 382 | 2.458 | 0.1304 | Yes | ||
| 4 | PAFAH1B1 | 513 | 2.259 | 0.1636 | Yes | ||
| 5 | CEP57 | 517 | 2.254 | 0.2024 | Yes | ||
| 6 | CKAP5 | 664 | 2.094 | 0.2320 | Yes | ||
| 7 | SFI1 | 682 | 2.078 | 0.2671 | Yes | ||
| 8 | PLK4 | 1193 | 1.711 | 0.2734 | Yes | ||
| 9 | CEP78 | 1386 | 1.610 | 0.2925 | Yes | ||
| 10 | YWHAE | 1550 | 1.540 | 0.3117 | Yes | ||
| 11 | SDCCAG8 | 1998 | 1.361 | 0.3149 | Yes | ||
| 12 | CEP110 | 2197 | 1.296 | 0.3282 | Yes | ||
| 13 | CDC25B | 2332 | 1.248 | 0.3437 | Yes | ||
| 14 | CEP135 | 2532 | 1.190 | 0.3552 | Yes | ||
| 15 | CLASP1 | 2741 | 1.140 | 0.3654 | Yes | ||
| 16 | MAPRE1 | 2852 | 1.117 | 0.3797 | Yes | ||
| 17 | PRKAR2B | 3025 | 1.077 | 0.3905 | Yes | ||
| 18 | CCNB1 | 3123 | 1.054 | 0.4043 | Yes | ||
| 19 | OFD1 | 3508 | 0.966 | 0.4035 | Yes | ||
| 20 | HSP90AA1 | 3539 | 0.961 | 0.4187 | Yes | ||
| 21 | PLK1 | 5036 | 0.703 | 0.3626 | No | ||
| 22 | SSNA1 | 5251 | 0.673 | 0.3644 | No | ||
| 23 | CCNA2 | 6116 | 0.542 | 0.3344 | No | ||
| 24 | TUBB2C | 6667 | 0.464 | 0.3173 | No | ||
| 25 | PPP2R1A | 7099 | 0.407 | 0.3046 | No | ||
| 26 | FGFR1OP | 7426 | 0.364 | 0.2960 | No | ||
| 27 | XPO1 | 7628 | 0.335 | 0.2927 | No | ||
| 28 | TUBG1 | 8749 | 0.184 | 0.2447 | No | ||
| 29 | TUBB4 | 8802 | 0.175 | 0.2453 | No | ||
| 30 | CCNB2 | 9421 | 0.086 | 0.2186 | No | ||
| 31 | DYNC1I2 | 9449 | 0.081 | 0.2188 | No | ||
| 32 | CCNA1 | 9609 | 0.059 | 0.2125 | No | ||
| 33 | MNAT1 | 9623 | 0.055 | 0.2129 | No | ||
| 34 | DCTN3 | 9769 | 0.033 | 0.2069 | No | ||
| 35 | CDC25C | 10914 | -0.143 | 0.1571 | No | ||
| 36 | CETN2 | 11494 | -0.216 | 0.1344 | No | ||
| 37 | CEP152 | 11626 | -0.231 | 0.1324 | No | ||
| 38 | DCTN2 | 12088 | -0.282 | 0.1162 | No | ||
| 39 | WEE1 | 12721 | -0.358 | 0.0935 | No | ||
| 40 | ACTR1A | 14012 | -0.510 | 0.0435 | No | ||
| 41 | PRKACA | 17631 | -0.996 | -0.1046 | No | ||
| 42 | CDK7 | 17893 | -1.041 | -0.0985 | No | ||
| 43 | CSNK1E | 18249 | -1.101 | -0.0956 | No | ||
| 44 | CDC25A | 18854 | -1.230 | -0.1019 | No | ||
| 45 | DYNLL1 | 18948 | -1.251 | -0.0845 | No | ||
| 46 | DYNC1H1 | 19637 | -1.435 | -0.0911 | No | ||
| 47 | PCNT | 20004 | -1.572 | -0.0806 | No | ||
| 48 | ODF2 | 20421 | -1.750 | -0.0694 | No | ||
| 49 | NUMA1 | 21077 | -2.170 | -0.0617 | No | ||
| 50 | CSNK1D | 21326 | -2.457 | -0.0306 | No | ||
| 51 | YWHAG | 21678 | -3.403 | 0.0123 | No |