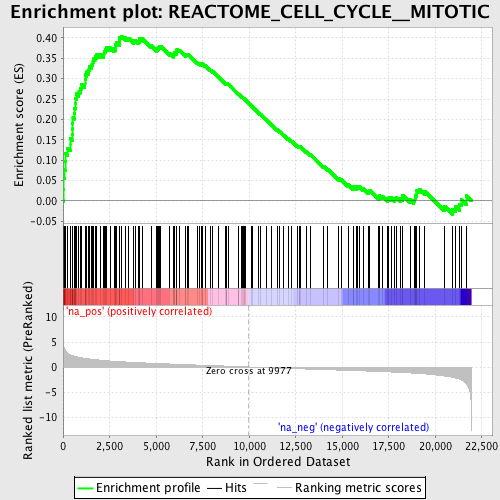
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | 0.40433222 |
| Normalized Enrichment Score (NES) | 1.9719187 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.049887225 |
| FWER p-Value | 0.19 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | CENPE | 21 | 4.670 | 0.0287 | Yes | ||
| 2 | ORC4L | 29 | 4.329 | 0.0558 | Yes | ||
| 3 | CDC27 | 73 | 3.632 | 0.0769 | Yes | ||
| 4 | SMC1A | 110 | 3.359 | 0.0965 | Yes | ||
| 5 | CCNH | 125 | 3.262 | 0.1165 | Yes | ||
| 6 | POLD3 | 240 | 2.785 | 0.1290 | Yes | ||
| 7 | NEK2 | 382 | 2.458 | 0.1381 | Yes | ||
| 8 | CLASP2 | 384 | 2.455 | 0.1536 | Yes | ||
| 9 | ORC1L | 505 | 2.267 | 0.1625 | Yes | ||
| 10 | PAFAH1B1 | 513 | 2.259 | 0.1765 | Yes | ||
| 11 | CEP57 | 517 | 2.254 | 0.1907 | Yes | ||
| 12 | TYMS | 530 | 2.236 | 0.2043 | Yes | ||
| 13 | RFC1 | 606 | 2.153 | 0.2145 | Yes | ||
| 14 | POLA1 | 621 | 2.138 | 0.2274 | Yes | ||
| 15 | CKAP5 | 664 | 2.094 | 0.2388 | Yes | ||
| 16 | SFI1 | 682 | 2.078 | 0.2512 | Yes | ||
| 17 | KIF23 | 717 | 2.037 | 0.2625 | Yes | ||
| 18 | RB1 | 852 | 1.918 | 0.2686 | Yes | ||
| 19 | CENPA | 937 | 1.859 | 0.2765 | Yes | ||
| 20 | DHFR | 983 | 1.831 | 0.2860 | Yes | ||
| 21 | PTTG1 | 1175 | 1.721 | 0.2882 | Yes | ||
| 22 | TAOK1 | 1176 | 1.720 | 0.2991 | Yes | ||
| 23 | PLK4 | 1193 | 1.711 | 0.3092 | Yes | ||
| 24 | RANBP2 | 1268 | 1.670 | 0.3164 | Yes | ||
| 25 | CEP78 | 1386 | 1.610 | 0.3213 | Yes | ||
| 26 | MCM3 | 1409 | 1.600 | 0.3304 | Yes | ||
| 27 | YWHAE | 1550 | 1.540 | 0.3337 | Yes | ||
| 28 | PSMC6 | 1604 | 1.520 | 0.3409 | Yes | ||
| 29 | PSMA6 | 1651 | 1.499 | 0.3483 | Yes | ||
| 30 | ORC2L | 1715 | 1.475 | 0.3548 | Yes | ||
| 31 | CENPC1 | 1816 | 1.429 | 0.3593 | Yes | ||
| 32 | SDCCAG8 | 1998 | 1.361 | 0.3596 | Yes | ||
| 33 | BUB3 | 2180 | 1.302 | 0.3596 | Yes | ||
| 34 | CEP110 | 2197 | 1.296 | 0.3671 | Yes | ||
| 35 | BUB1 | 2260 | 1.274 | 0.3723 | Yes | ||
| 36 | CDC25B | 2332 | 1.248 | 0.3770 | Yes | ||
| 37 | CEP135 | 2532 | 1.190 | 0.3754 | Yes | ||
| 38 | CLASP1 | 2741 | 1.140 | 0.3731 | Yes | ||
| 39 | CDC20 | 2828 | 1.122 | 0.3762 | Yes | ||
| 40 | PSMD9 | 2832 | 1.121 | 0.3832 | Yes | ||
| 41 | MAPRE1 | 2852 | 1.117 | 0.3894 | Yes | ||
| 42 | CDC45L | 3011 | 1.081 | 0.3890 | Yes | ||
| 43 | PRKAR2B | 3025 | 1.077 | 0.3952 | Yes | ||
| 44 | UBE2C | 3043 | 1.072 | 0.4013 | Yes | ||
| 45 | CCNB1 | 3123 | 1.054 | 0.4043 | Yes | ||
| 46 | PSMB2 | 3343 | 1.003 | 0.4006 | No | ||
| 47 | OFD1 | 3508 | 0.966 | 0.3992 | No | ||
| 48 | NUP160 | 3780 | 0.912 | 0.3926 | No | ||
| 49 | RPA1 | 3890 | 0.893 | 0.3933 | No | ||
| 50 | CKS1B | 4061 | 0.861 | 0.3909 | No | ||
| 51 | STAG2 | 4119 | 0.852 | 0.3937 | No | ||
| 52 | KNTC1 | 4123 | 0.851 | 0.3990 | No | ||
| 53 | PSMA1 | 4270 | 0.827 | 0.3975 | No | ||
| 54 | MCM5 | 4731 | 0.752 | 0.3812 | No | ||
| 55 | PLK1 | 5036 | 0.703 | 0.3717 | No | ||
| 56 | PSMD14 | 5093 | 0.696 | 0.3735 | No | ||
| 57 | SGOL1 | 5135 | 0.690 | 0.3760 | No | ||
| 58 | RFC5 | 5190 | 0.682 | 0.3779 | No | ||
| 59 | SSNA1 | 5251 | 0.673 | 0.3794 | No | ||
| 60 | POLE | 5732 | 0.602 | 0.3612 | No | ||
| 61 | MCM7 | 5913 | 0.572 | 0.3566 | No | ||
| 62 | MCM2 | 5944 | 0.567 | 0.3588 | No | ||
| 63 | FEN1 | 5963 | 0.565 | 0.3615 | No | ||
| 64 | KIF2A | 5971 | 0.564 | 0.3648 | No | ||
| 65 | PRIM1 | 6109 | 0.544 | 0.3620 | No | ||
| 66 | CUL1 | 6112 | 0.543 | 0.3653 | No | ||
| 67 | PMF1 | 6114 | 0.542 | 0.3687 | No | ||
| 68 | CCNA2 | 6116 | 0.542 | 0.3721 | No | ||
| 69 | CDKN1A | 6257 | 0.518 | 0.3690 | No | ||
| 70 | RFC2 | 6568 | 0.478 | 0.3578 | No | ||
| 71 | RAD21 | 6579 | 0.476 | 0.3603 | No | ||
| 72 | TUBB2C | 6667 | 0.464 | 0.3593 | No | ||
| 73 | RPA2 | 6728 | 0.456 | 0.3594 | No | ||
| 74 | MCM4 | 7237 | 0.389 | 0.3386 | No | ||
| 75 | POLD1 | 7306 | 0.379 | 0.3379 | No | ||
| 76 | FGFR1OP | 7426 | 0.364 | 0.3347 | No | ||
| 77 | E2F1 | 7467 | 0.357 | 0.3351 | No | ||
| 78 | POLA2 | 7492 | 0.354 | 0.3363 | No | ||
| 79 | XPO1 | 7628 | 0.335 | 0.3322 | No | ||
| 80 | CDK4 | 7918 | 0.298 | 0.3209 | No | ||
| 81 | PSMD12 | 8046 | 0.281 | 0.3168 | No | ||
| 82 | POLD2 | 8333 | 0.241 | 0.3052 | No | ||
| 83 | TUBG1 | 8749 | 0.184 | 0.2873 | No | ||
| 84 | MAD2L1 | 8752 | 0.183 | 0.2884 | No | ||
| 85 | RFC4 | 8786 | 0.177 | 0.2880 | No | ||
| 86 | TUBB4 | 8802 | 0.175 | 0.2884 | No | ||
| 87 | PSMB4 | 8878 | 0.165 | 0.2860 | No | ||
| 88 | NUP37 | 9417 | 0.087 | 0.2619 | No | ||
| 89 | CCNB2 | 9421 | 0.086 | 0.2623 | No | ||
| 90 | DYNC1I2 | 9449 | 0.081 | 0.2616 | No | ||
| 91 | CCNA1 | 9609 | 0.059 | 0.2547 | No | ||
| 92 | MNAT1 | 9623 | 0.055 | 0.2544 | No | ||
| 93 | SGOL2 | 9687 | 0.046 | 0.2518 | No | ||
| 94 | PSMA4 | 9724 | 0.040 | 0.2504 | No | ||
| 95 | DCTN3 | 9769 | 0.033 | 0.2486 | No | ||
| 96 | PCNA | 9779 | 0.031 | 0.2484 | No | ||
| 97 | PSMD3 | 10136 | -0.024 | 0.2322 | No | ||
| 98 | RPA3 | 10199 | -0.033 | 0.2296 | No | ||
| 99 | PSMD8 | 10484 | -0.076 | 0.2171 | No | ||
| 100 | BIRC5 | 10590 | -0.093 | 0.2128 | No | ||
| 101 | CDC25C | 10914 | -0.143 | 0.1989 | No | ||
| 102 | PSMA3 | 11218 | -0.184 | 0.1862 | No | ||
| 103 | CETN2 | 11494 | -0.216 | 0.1749 | No | ||
| 104 | CEP152 | 11626 | -0.231 | 0.1704 | No | ||
| 105 | ZW10 | 11856 | -0.258 | 0.1615 | No | ||
| 106 | DCTN2 | 12088 | -0.282 | 0.1527 | No | ||
| 107 | PSMC4 | 12263 | -0.303 | 0.1466 | No | ||
| 108 | BUB1B | 12571 | -0.341 | 0.1347 | No | ||
| 109 | PSMB9 | 12679 | -0.353 | 0.1320 | No | ||
| 110 | WEE1 | 12721 | -0.358 | 0.1324 | No | ||
| 111 | CCNE1 | 12750 | -0.362 | 0.1334 | No | ||
| 112 | NUP107 | 13083 | -0.401 | 0.1208 | No | ||
| 113 | PSMD6 | 13284 | -0.423 | 0.1143 | No | ||
| 114 | ACTR1A | 14012 | -0.510 | 0.0841 | No | ||
| 115 | KIF20A | 14219 | -0.535 | 0.0781 | No | ||
| 116 | PSMB5 | 14806 | -0.607 | 0.0551 | No | ||
| 117 | GMNN | 14935 | -0.622 | 0.0531 | No | ||
| 118 | CDC16 | 15314 | -0.667 | 0.0400 | No | ||
| 119 | LIG1 | 15585 | -0.699 | 0.0321 | No | ||
| 120 | PSMC1 | 15623 | -0.705 | 0.0348 | No | ||
| 121 | ORC5L | 15771 | -0.724 | 0.0327 | No | ||
| 122 | RANGAP1 | 15793 | -0.727 | 0.0363 | No | ||
| 123 | PSMD4 | 15917 | -0.744 | 0.0354 | No | ||
| 124 | PSMD5 | 16120 | -0.773 | 0.0310 | No | ||
| 125 | PSMC5 | 16387 | -0.809 | 0.0240 | No | ||
| 126 | RFC3 | 16485 | -0.821 | 0.0247 | No | ||
| 127 | PSMD11 | 16972 | -0.887 | 0.0081 | No | ||
| 128 | PSMA5 | 16974 | -0.887 | 0.0136 | No | ||
| 129 | NUP43 | 17142 | -0.914 | 0.0118 | No | ||
| 130 | PSMC3 | 17413 | -0.955 | 0.0055 | No | ||
| 131 | POLE2 | 17509 | -0.973 | 0.0073 | No | ||
| 132 | PRKACA | 17631 | -0.996 | 0.0080 | No | ||
| 133 | TK2 | 17818 | -1.025 | 0.0060 | No | ||
| 134 | CDK7 | 17893 | -1.041 | 0.0092 | No | ||
| 135 | PSMB10 | 18105 | -1.077 | 0.0064 | No | ||
| 136 | PSMA2 | 18226 | -1.098 | 0.0078 | No | ||
| 137 | CSNK1E | 18249 | -1.101 | 0.0138 | No | ||
| 138 | PSMA7 | 18640 | -1.183 | 0.0034 | No | ||
| 139 | CDC25A | 18854 | -1.230 | 0.0014 | No | ||
| 140 | PSMD7 | 18907 | -1.242 | 0.0069 | No | ||
| 141 | DYNLL1 | 18948 | -1.251 | 0.0130 | No | ||
| 142 | MCM10 | 19005 | -1.263 | 0.0184 | No | ||
| 143 | CDK2 | 19007 | -1.263 | 0.0264 | No | ||
| 144 | PPP1CC | 19133 | -1.291 | 0.0289 | No | ||
| 145 | PSMD2 | 19433 | -1.375 | 0.0239 | No | ||
| 146 | PSME3 | 20480 | -1.781 | -0.0128 | No | ||
| 147 | PSMB1 | 20928 | -2.055 | -0.0203 | No | ||
| 148 | NUMA1 | 21077 | -2.170 | -0.0133 | No | ||
| 149 | UBE2D1 | 21292 | -2.403 | -0.0079 | No | ||
| 150 | ZWINT | 21393 | -2.564 | 0.0038 | No | ||
| 151 | YWHAG | 21678 | -3.403 | 0.0123 | No |
