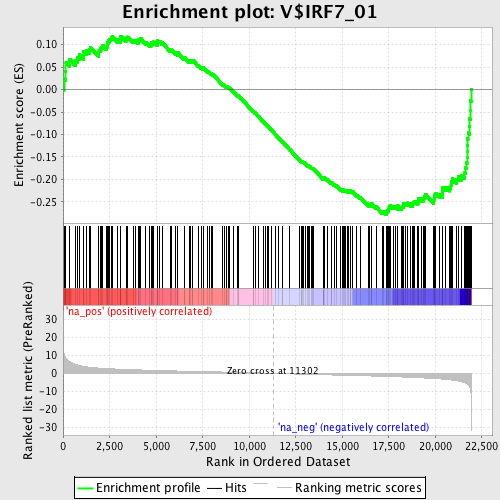
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_SCID |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$IRF7_01 |
| Enrichment Score (ES) | -0.27813032 |
| Normalized Enrichment Score (NES) | -1.3737707 |
| Nominal p-value | 0.0049751243 |
| FDR q-value | 0.3277949 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | YTHDF2 | 48 | 11.012 | 0.0243 | No | ||
| 2 | KPNA3 | 138 | 8.635 | 0.0410 | No | ||
| 3 | EPSTI1 | 153 | 8.473 | 0.0608 | No | ||
| 4 | COMMD10 | 345 | 6.508 | 0.0677 | No | ||
| 5 | AGPAT4 | 649 | 4.944 | 0.0657 | No | ||
| 6 | CUL2 | 773 | 4.616 | 0.0712 | No | ||
| 7 | TLK2 | 861 | 4.415 | 0.0778 | No | ||
| 8 | DICER1 | 1101 | 3.888 | 0.0762 | No | ||
| 9 | E2F3 | 1108 | 3.880 | 0.0852 | No | ||
| 10 | PPP2R2A | 1272 | 3.605 | 0.0864 | No | ||
| 11 | STX11 | 1402 | 3.428 | 0.0888 | No | ||
| 12 | RBL1 | 1464 | 3.353 | 0.0940 | No | ||
| 13 | PSCD3 | 1893 | 2.943 | 0.0815 | No | ||
| 14 | FLRT2 | 1927 | 2.918 | 0.0870 | No | ||
| 15 | SOX4 | 1998 | 2.850 | 0.0906 | No | ||
| 16 | TRPM3 | 2056 | 2.804 | 0.0948 | No | ||
| 17 | NPR3 | 2136 | 2.742 | 0.0977 | No | ||
| 18 | LSM6 | 2345 | 2.621 | 0.0945 | No | ||
| 19 | IFIT2 | 2363 | 2.607 | 0.1000 | No | ||
| 20 | ANGPTL4 | 2380 | 2.598 | 0.1055 | No | ||
| 21 | HTR5A | 2442 | 2.565 | 0.1089 | No | ||
| 22 | SIPA1L1 | 2512 | 2.532 | 0.1118 | No | ||
| 23 | EGR4 | 2573 | 2.496 | 0.1151 | No | ||
| 24 | NOVA2 | 2645 | 2.458 | 0.1177 | No | ||
| 25 | EYA1 | 2914 | 2.335 | 0.1110 | No | ||
| 26 | RASGEF1A | 3061 | 2.274 | 0.1098 | No | ||
| 27 | MRPS18B | 3068 | 2.271 | 0.1150 | No | ||
| 28 | PSMA3 | 3100 | 2.259 | 0.1190 | No | ||
| 29 | ELAVL2 | 3380 | 2.149 | 0.1114 | No | ||
| 30 | RAPH1 | 3416 | 2.135 | 0.1149 | No | ||
| 31 | CPT1A | 3480 | 2.111 | 0.1171 | No | ||
| 32 | EPHA7 | 3763 | 2.004 | 0.1090 | No | ||
| 33 | SOX5 | 3874 | 1.966 | 0.1087 | No | ||
| 34 | CACNA2D3 | 4027 | 1.917 | 0.1063 | No | ||
| 35 | OPTN | 4033 | 1.915 | 0.1107 | No | ||
| 36 | TAF5 | 4094 | 1.897 | 0.1125 | No | ||
| 37 | A2BP1 | 4149 | 1.881 | 0.1145 | No | ||
| 38 | GRM7 | 4452 | 1.782 | 0.1049 | No | ||
| 39 | HS6ST2 | 4643 | 1.715 | 0.1003 | No | ||
| 40 | IL18BP | 4724 | 1.690 | 0.1007 | No | ||
| 41 | SALL3 | 4726 | 1.690 | 0.1048 | No | ||
| 42 | CACNA1A | 4812 | 1.664 | 0.1049 | No | ||
| 43 | CDYL | 4873 | 1.645 | 0.1061 | No | ||
| 44 | HMGB3 | 5053 | 1.589 | 0.1017 | No | ||
| 45 | LAPTM4A | 5059 | 1.587 | 0.1053 | No | ||
| 46 | MSX1 | 5068 | 1.584 | 0.1087 | No | ||
| 47 | PRDM16 | 5169 | 1.552 | 0.1079 | No | ||
| 48 | NOG | 5328 | 1.510 | 0.1042 | No | ||
| 49 | LHX9 | 5748 | 1.396 | 0.0883 | No | ||
| 50 | GATA6 | 5819 | 1.378 | 0.0884 | No | ||
| 51 | TAPBP | 6035 | 1.327 | 0.0818 | No | ||
| 52 | JMJD1A | 6161 | 1.293 | 0.0791 | No | ||
| 53 | CXCL16 | 6167 | 1.291 | 0.0820 | No | ||
| 54 | ERG | 6504 | 1.216 | 0.0695 | No | ||
| 55 | LIF | 6518 | 1.214 | 0.0718 | No | ||
| 56 | ZMYND15 | 6770 | 1.155 | 0.0631 | No | ||
| 57 | GDNF | 6792 | 1.149 | 0.0649 | No | ||
| 58 | ELK4 | 6845 | 1.135 | 0.0652 | No | ||
| 59 | ECEL1 | 6945 | 1.112 | 0.0633 | No | ||
| 60 | DMD | 6960 | 1.109 | 0.0654 | No | ||
| 61 | PITX2 | 7285 | 1.032 | 0.0530 | No | ||
| 62 | PCBP1 | 7434 | 0.998 | 0.0486 | No | ||
| 63 | ARTN | 7520 | 0.983 | 0.0470 | No | ||
| 64 | APOBEC2 | 7524 | 0.982 | 0.0493 | No | ||
| 65 | LDB2 | 7757 | 0.929 | 0.0408 | No | ||
| 66 | SST | 7846 | 0.908 | 0.0390 | No | ||
| 67 | USP2 | 7960 | 0.881 | 0.0359 | No | ||
| 68 | KIRREL3 | 8051 | 0.858 | 0.0338 | No | ||
| 69 | FOS | 8574 | 0.740 | 0.0116 | No | ||
| 70 | LGALS3BP | 8647 | 0.726 | 0.0101 | No | ||
| 71 | RDH10 | 8761 | 0.696 | 0.0066 | No | ||
| 72 | HOXD13 | 8788 | 0.690 | 0.0070 | No | ||
| 73 | LOX | 8882 | 0.666 | 0.0044 | No | ||
| 74 | CXCL10 | 8931 | 0.654 | 0.0037 | No | ||
| 75 | ZBTB10 | 9144 | 0.605 | -0.0046 | No | ||
| 76 | PKN3 | 9359 | 0.557 | -0.0130 | No | ||
| 77 | DDX58 | 9403 | 0.547 | -0.0137 | No | ||
| 78 | HOXC10 | 9411 | 0.544 | -0.0127 | No | ||
| 79 | MAP3K8 | 10208 | 0.325 | -0.0485 | No | ||
| 80 | NMNAT1 | 10210 | 0.324 | -0.0478 | No | ||
| 81 | ZHX2 | 10314 | 0.296 | -0.0518 | No | ||
| 82 | PDGFC | 10505 | 0.239 | -0.0600 | No | ||
| 83 | EN1 | 10768 | 0.162 | -0.0716 | No | ||
| 84 | IFNB1 | 10886 | 0.122 | -0.0767 | No | ||
| 85 | PLEK2 | 10983 | 0.096 | -0.0809 | No | ||
| 86 | CXCR4 | 11032 | 0.079 | -0.0829 | No | ||
| 87 | JPH4 | 11177 | 0.037 | -0.0894 | No | ||
| 88 | GNGT2 | 11430 | -0.045 | -0.1009 | No | ||
| 89 | SLC4A10 | 11574 | -0.090 | -0.1072 | No | ||
| 90 | ERBB2 | 11776 | -0.149 | -0.1161 | No | ||
| 91 | GSC | 11814 | -0.162 | -0.1174 | No | ||
| 92 | FIBCD1 | 12190 | -0.272 | -0.1340 | No | ||
| 93 | ARRDC3 | 12703 | -0.422 | -0.1565 | No | ||
| 94 | PSME1 | 12817 | -0.453 | -0.1606 | No | ||
| 95 | STX16 | 12833 | -0.458 | -0.1602 | No | ||
| 96 | TCF15 | 12861 | -0.465 | -0.1603 | No | ||
| 97 | ITGB7 | 12915 | -0.481 | -0.1616 | No | ||
| 98 | NT5C3 | 13048 | -0.518 | -0.1664 | No | ||
| 99 | ZFP36L1 | 13142 | -0.543 | -0.1694 | No | ||
| 100 | EGFL6 | 13173 | -0.553 | -0.1694 | No | ||
| 101 | LHX5 | 13255 | -0.575 | -0.1718 | No | ||
| 102 | LRRTM4 | 13357 | -0.607 | -0.1750 | No | ||
| 103 | CCL3 | 13385 | -0.619 | -0.1747 | No | ||
| 104 | RFX5 | 13427 | -0.629 | -0.1751 | No | ||
| 105 | TREX2 | 13981 | -0.774 | -0.1986 | No | ||
| 106 | DLX4 | 13986 | -0.774 | -0.1969 | No | ||
| 107 | HOXA10 | 13989 | -0.775 | -0.1952 | No | ||
| 108 | AAMP | 14068 | -0.796 | -0.1968 | No | ||
| 109 | OSM | 14179 | -0.824 | -0.1999 | No | ||
| 110 | TLX2 | 14397 | -0.877 | -0.2078 | No | ||
| 111 | TUBD1 | 14426 | -0.885 | -0.2069 | No | ||
| 112 | BMP5 | 14567 | -0.923 | -0.2111 | No | ||
| 113 | FEV | 14678 | -0.953 | -0.2139 | No | ||
| 114 | BNC2 | 14908 | -1.020 | -0.2220 | No | ||
| 115 | POLL | 15016 | -1.051 | -0.2243 | No | ||
| 116 | HIVEP1 | 15050 | -1.060 | -0.2233 | No | ||
| 117 | MPL | 15121 | -1.079 | -0.2239 | No | ||
| 118 | NFIX | 15179 | -1.096 | -0.2239 | No | ||
| 119 | CHST8 | 15281 | -1.123 | -0.2258 | No | ||
| 120 | BST2 | 15313 | -1.131 | -0.2245 | No | ||
| 121 | RFX4 | 15426 | -1.162 | -0.2269 | No | ||
| 122 | NOS1 | 15450 | -1.168 | -0.2251 | No | ||
| 123 | AMY2A | 15565 | -1.197 | -0.2275 | No | ||
| 124 | HOXB8 | 15788 | -1.257 | -0.2347 | No | ||
| 125 | GPX1 | 15966 | -1.302 | -0.2397 | No | ||
| 126 | COL12A1 | 16397 | -1.414 | -0.2560 | No | ||
| 127 | PDE1B | 16445 | -1.428 | -0.2547 | No | ||
| 128 | VIT | 16555 | -1.455 | -0.2562 | No | ||
| 129 | PDE4D | 16569 | -1.458 | -0.2533 | No | ||
| 130 | GRM3 | 16820 | -1.532 | -0.2611 | No | ||
| 131 | ATP5G2 | 17156 | -1.634 | -0.2726 | No | ||
| 132 | FHL3 | 17200 | -1.648 | -0.2706 | No | ||
| 133 | DLL4 | 17365 | -1.707 | -0.2740 | Yes | ||
| 134 | BAT5 | 17386 | -1.715 | -0.2708 | Yes | ||
| 135 | USP18 | 17436 | -1.730 | -0.2689 | Yes | ||
| 136 | BCL6 | 17488 | -1.746 | -0.2670 | Yes | ||
| 137 | KIF13A | 17508 | -1.753 | -0.2637 | Yes | ||
| 138 | UPF2 | 17531 | -1.761 | -0.2604 | Yes | ||
| 139 | DLEU2 | 17567 | -1.775 | -0.2578 | Yes | ||
| 140 | FOXP2 | 17726 | -1.828 | -0.2606 | Yes | ||
| 141 | LHX6 | 17837 | -1.865 | -0.2612 | Yes | ||
| 142 | POPDC2 | 17959 | -1.905 | -0.2622 | Yes | ||
| 143 | MOV10 | 17967 | -1.908 | -0.2579 | Yes | ||
| 144 | NRP1 | 18183 | -1.990 | -0.2630 | Yes | ||
| 145 | PLXNC1 | 18210 | -2.000 | -0.2594 | Yes | ||
| 146 | PIK3CD | 18281 | -2.027 | -0.2577 | Yes | ||
| 147 | HOXB3 | 18298 | -2.036 | -0.2535 | Yes | ||
| 148 | UBE1L | 18393 | -2.076 | -0.2528 | Yes | ||
| 149 | PPP1R10 | 18497 | -2.120 | -0.2525 | Yes | ||
| 150 | RAPGEF6 | 18652 | -2.185 | -0.2543 | Yes | ||
| 151 | IL4 | 18791 | -2.244 | -0.2552 | Yes | ||
| 152 | DDX17 | 18811 | -2.253 | -0.2507 | Yes | ||
| 153 | NR4A2 | 18880 | -2.284 | -0.2483 | Yes | ||
| 154 | ESR1 | 19068 | -2.374 | -0.2512 | Yes | ||
| 155 | VPS24 | 19111 | -2.397 | -0.2473 | Yes | ||
| 156 | CCDC6 | 19113 | -2.398 | -0.2416 | Yes | ||
| 157 | LZIC | 19261 | -2.479 | -0.2424 | Yes | ||
| 158 | SLC25A28 | 19390 | -2.542 | -0.2421 | Yes | ||
| 159 | RFX3 | 19424 | -2.560 | -0.2375 | Yes | ||
| 160 | MAP2K5 | 19463 | -2.584 | -0.2330 | Yes | ||
| 161 | COL11A2 | 19914 | -2.838 | -0.2468 | Yes | ||
| 162 | TAP1 | 19931 | -2.847 | -0.2407 | Yes | ||
| 163 | ABI3 | 19951 | -2.857 | -0.2347 | Yes | ||
| 164 | CALD1 | 20032 | -2.916 | -0.2314 | Yes | ||
| 165 | ADAR | 20231 | -3.048 | -0.2331 | Yes | ||
| 166 | EVL | 20375 | -3.154 | -0.2321 | Yes | ||
| 167 | SOCS1 | 20394 | -3.165 | -0.2253 | Yes | ||
| 168 | RPS6KB1 | 20405 | -3.174 | -0.2181 | Yes | ||
| 169 | MAP2K6 | 20569 | -3.327 | -0.2176 | Yes | ||
| 170 | NR4A3 | 20751 | -3.509 | -0.2174 | Yes | ||
| 171 | DAPP1 | 20841 | -3.613 | -0.2128 | Yes | ||
| 172 | LRP6 | 20873 | -3.655 | -0.2054 | Yes | ||
| 173 | BRUNOL4 | 20915 | -3.703 | -0.1984 | Yes | ||
| 174 | DLX1 | 21159 | -4.071 | -0.1998 | Yes | ||
| 175 | ARF6 | 21229 | -4.197 | -0.1928 | Yes | ||
| 176 | ZDHHC14 | 21422 | -4.658 | -0.1904 | Yes | ||
| 177 | PRKCBP1 | 21590 | -5.204 | -0.1856 | Yes | ||
| 178 | FLI1 | 21627 | -5.352 | -0.1743 | Yes | ||
| 179 | SLC12A7 | 21674 | -5.542 | -0.1631 | Yes | ||
| 180 | GDI1 | 21704 | -5.680 | -0.1507 | Yes | ||
| 181 | MXD3 | 21708 | -5.712 | -0.1371 | Yes | ||
| 182 | GNA13 | 21713 | -5.757 | -0.1234 | Yes | ||
| 183 | TSC1 | 21728 | -5.921 | -0.1098 | Yes | ||
| 184 | TIA1 | 21757 | -6.125 | -0.0963 | Yes | ||
| 185 | CBX4 | 21833 | -7.190 | -0.0825 | Yes | ||
| 186 | IRF2 | 21846 | -7.452 | -0.0651 | Yes | ||
| 187 | TCF12 | 21878 | -8.562 | -0.0459 | Yes | ||
| 188 | PSMB9 | 21883 | -8.701 | -0.0251 | Yes | ||
| 189 | CASP7 | 21924 | -11.616 | 0.0011 | Yes |
