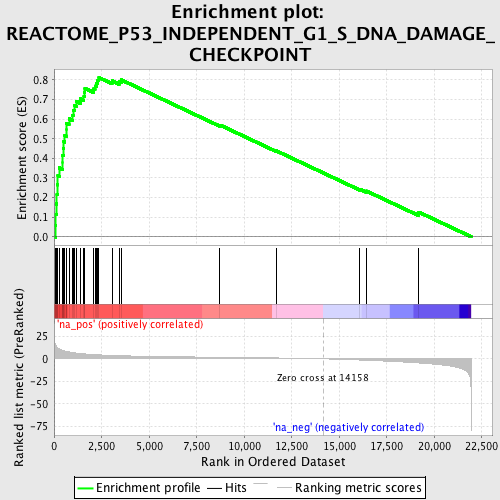
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set01_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | REACTOME_P53_INDEPENDENT_G1_S_DNA_DAMAGE_CHECKPOINT |
| Enrichment Score (ES) | 0.8118712 |
| Normalized Enrichment Score (NES) | 2.8547728 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMC2 | 77 | 15.326 | 0.0568 | Yes | ||
| 2 | PSMB2 | 97 | 14.717 | 0.1138 | Yes | ||
| 3 | PSMB1 | 121 | 13.977 | 0.1678 | Yes | ||
| 4 | PSMA7 | 149 | 13.085 | 0.2180 | Yes | ||
| 5 | PSMB3 | 190 | 12.244 | 0.2644 | Yes | ||
| 6 | PSMD14 | 200 | 11.991 | 0.3112 | Yes | ||
| 7 | PSMA2 | 273 | 11.079 | 0.3515 | Yes | ||
| 8 | PSMD7 | 453 | 9.308 | 0.3799 | Yes | ||
| 9 | PSMA5 | 455 | 9.295 | 0.4165 | Yes | ||
| 10 | PSMD12 | 484 | 9.145 | 0.4512 | Yes | ||
| 11 | PSMC6 | 512 | 8.965 | 0.4852 | Yes | ||
| 12 | PSMB9 | 563 | 8.611 | 0.5168 | Yes | ||
| 13 | PSMB5 | 639 | 8.261 | 0.5459 | Yes | ||
| 14 | PSMB6 | 642 | 8.237 | 0.5782 | Yes | ||
| 15 | PSMC5 | 787 | 7.643 | 0.6017 | Yes | ||
| 16 | PSMB4 | 968 | 6.973 | 0.6210 | Yes | ||
| 17 | PSMA1 | 1002 | 6.871 | 0.6465 | Yes | ||
| 18 | PSMA6 | 1086 | 6.599 | 0.6687 | Yes | ||
| 19 | PSMA4 | 1169 | 6.376 | 0.6900 | Yes | ||
| 20 | PSMD9 | 1379 | 5.837 | 0.7034 | Yes | ||
| 21 | PSMB10 | 1569 | 5.467 | 0.7163 | Yes | ||
| 22 | PSMD5 | 1587 | 5.429 | 0.7369 | Yes | ||
| 23 | PSMD3 | 1622 | 5.364 | 0.7565 | Yes | ||
| 24 | PSMC3 | 2088 | 4.711 | 0.7538 | Yes | ||
| 25 | PSME3 | 2153 | 4.608 | 0.7690 | Yes | ||
| 26 | PSMA3 | 2238 | 4.484 | 0.7828 | Yes | ||
| 27 | PSMD8 | 2299 | 4.418 | 0.7975 | Yes | ||
| 28 | CDC25A | 2359 | 4.348 | 0.8119 | Yes | ||
| 29 | PSMC4 | 3050 | 3.765 | 0.7952 | No | ||
| 30 | PSMD6 | 3419 | 3.480 | 0.7921 | No | ||
| 31 | PSMC1 | 3537 | 3.415 | 0.8002 | No | ||
| 32 | PSME1 | 8729 | 1.685 | 0.5699 | No | ||
| 33 | PSMD2 | 11699 | 0.938 | 0.4381 | No | ||
| 34 | PSMD4 | 16083 | -1.310 | 0.2432 | No | ||
| 35 | PSMD10 | 16435 | -1.601 | 0.2335 | No | ||
| 36 | PSMD11 | 19193 | -4.572 | 0.1257 | No |
