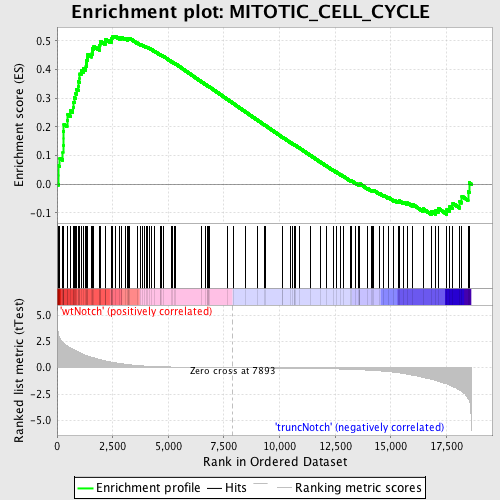
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | MITOTIC_CELL_CYCLE |
| Enrichment Score (ES) | 0.5169219 |
| Normalized Enrichment Score (NES) | 1.5830983 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.31623277 |
| FWER p-Value | 0.99 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NCAPH | 6220435 | 56 | 3.183 | 0.0310 | Yes | ||
| 2 | AKAP8 | 4210026 | 57 | 3.171 | 0.0649 | Yes | ||
| 3 | KPNA2 | 3450114 | 125 | 2.781 | 0.0911 | Yes | ||
| 4 | BUB1B | 1450288 | 261 | 2.413 | 0.1096 | Yes | ||
| 5 | POLE | 6020538 | 265 | 2.398 | 0.1350 | Yes | ||
| 6 | ANAPC4 | 4540338 | 272 | 2.384 | 0.1602 | Yes | ||
| 7 | MAD2L1 | 4480725 | 273 | 2.379 | 0.1857 | Yes | ||
| 8 | TPX2 | 6420324 | 306 | 2.306 | 0.2086 | Yes | ||
| 9 | CUL3 | 1850520 | 446 | 2.066 | 0.2232 | Yes | ||
| 10 | SKP2 | 360711 380093 4810368 | 483 | 2.012 | 0.2427 | Yes | ||
| 11 | ANAPC5 | 730164 7100685 | 597 | 1.862 | 0.2565 | Yes | ||
| 12 | NUSAP1 | 940048 3120435 | 714 | 1.733 | 0.2688 | Yes | ||
| 13 | CDC7 | 4060546 4850041 | 725 | 1.718 | 0.2866 | Yes | ||
| 14 | CUL1 | 1990632 | 772 | 1.677 | 0.3021 | Yes | ||
| 15 | PPP6C | 2450670 | 840 | 1.613 | 0.3157 | Yes | ||
| 16 | NDC80 | 4120465 | 867 | 1.583 | 0.3312 | Yes | ||
| 17 | ZWINT | 6940670 | 954 | 1.481 | 0.3424 | Yes | ||
| 18 | PRC1 | 870092 5890204 | 962 | 1.472 | 0.3578 | Yes | ||
| 19 | KIF23 | 5570112 | 1019 | 1.409 | 0.3698 | Yes | ||
| 20 | GSPT1 | 5420050 | 1023 | 1.408 | 0.3847 | Yes | ||
| 21 | CDKN1C | 6520577 | 1081 | 1.353 | 0.3961 | Yes | ||
| 22 | MPHOSPH6 | 5570164 | 1179 | 1.259 | 0.4043 | Yes | ||
| 23 | KHDRBS1 | 1240403 6040040 | 1297 | 1.152 | 0.4103 | Yes | ||
| 24 | AURKA | 780537 | 1317 | 1.140 | 0.4215 | Yes | ||
| 25 | SMC1A | 3060600 5700148 5890113 6370154 | 1333 | 1.130 | 0.4327 | Yes | ||
| 26 | ZW10 | 2900735 3520687 | 1362 | 1.103 | 0.4430 | Yes | ||
| 27 | CDKN2A | 4670215 4760047 | 1380 | 1.090 | 0.4538 | Yes | ||
| 28 | PCBP4 | 6350722 | 1563 | 0.991 | 0.4545 | Yes | ||
| 29 | CD28 | 1400739 4210093 | 1578 | 0.983 | 0.4643 | Yes | ||
| 30 | CDC16 | 1940706 | 1589 | 0.980 | 0.4742 | Yes | ||
| 31 | KNTC1 | 430079 | 1652 | 0.940 | 0.4809 | Yes | ||
| 32 | BIRC5 | 110408 580014 1770632 | 1894 | 0.790 | 0.4763 | Yes | ||
| 33 | ABL1 | 1050593 2030050 4010114 | 1898 | 0.789 | 0.4846 | Yes | ||
| 34 | CDK2AP1 | 2340156 | 1928 | 0.770 | 0.4913 | Yes | ||
| 35 | DLG7 | 3120041 | 1942 | 0.763 | 0.4987 | Yes | ||
| 36 | CDC6 | 4570296 5360600 | 2159 | 0.641 | 0.4939 | Yes | ||
| 37 | STMN1 | 1990717 | 2167 | 0.636 | 0.5003 | Yes | ||
| 38 | CCNA2 | 5290075 | 2176 | 0.634 | 0.5067 | Yes | ||
| 39 | SUGT1 | 1690670 | 2430 | 0.532 | 0.4987 | Yes | ||
| 40 | CIT | 2370601 | 2435 | 0.528 | 0.5041 | Yes | ||
| 41 | SMC4 | 5910240 | 2448 | 0.524 | 0.5091 | Yes | ||
| 42 | EGF | 5220154 | 2474 | 0.515 | 0.5132 | Yes | ||
| 43 | CUL4A | 1170088 2320008 2470278 | 2506 | 0.504 | 0.5169 | Yes | ||
| 44 | SSSCA1 | 1240402 780309 | 2608 | 0.469 | 0.5165 | No | ||
| 45 | PPP5C | 3130047 | 2795 | 0.409 | 0.5108 | No | ||
| 46 | NOLC1 | 2350195 | 2875 | 0.381 | 0.5106 | No | ||
| 47 | GFI1 | 730180 | 2907 | 0.371 | 0.5129 | No | ||
| 48 | KIF22 | 1190368 | 3067 | 0.321 | 0.5077 | No | ||
| 49 | LATS2 | 4480593 6020494 | 3166 | 0.294 | 0.5056 | No | ||
| 50 | RAD17 | 5220739 | 3194 | 0.288 | 0.5072 | No | ||
| 51 | CDK2 | 130484 2260301 4010088 5050110 | 3216 | 0.284 | 0.5091 | No | ||
| 52 | ACVR1B | 3610446 5570195 | 3271 | 0.272 | 0.5091 | No | ||
| 53 | GFI1B | 4570131 | 3607 | 0.204 | 0.4931 | No | ||
| 54 | PIN1 | 2970408 | 3758 | 0.180 | 0.4869 | No | ||
| 55 | TGFA | 1570332 | 3820 | 0.172 | 0.4855 | No | ||
| 56 | SMC3 | 870546 1400411 5700039 6450577 | 3910 | 0.158 | 0.4823 | No | ||
| 57 | NPM2 | 6040139 | 4003 | 0.148 | 0.4789 | No | ||
| 58 | CDKN1B | 3800025 6450044 | 4044 | 0.142 | 0.4783 | No | ||
| 59 | DDX11 | 6590671 | 4135 | 0.133 | 0.4749 | No | ||
| 60 | CDC25C | 2570673 4760161 6520707 | 4241 | 0.124 | 0.4705 | No | ||
| 61 | USH1C | 1570372 4230364 | 4394 | 0.110 | 0.4635 | No | ||
| 62 | PML | 50093 2190435 2450402 3840082 | 4633 | 0.093 | 0.4516 | No | ||
| 63 | FBXO5 | 2630551 | 4653 | 0.092 | 0.4515 | No | ||
| 64 | ATM | 3610110 4050524 | 4686 | 0.090 | 0.4508 | No | ||
| 65 | DCTN3 | 6840131 | 4767 | 0.085 | 0.4474 | No | ||
| 66 | CDC23 | 3190593 | 5157 | 0.066 | 0.4270 | No | ||
| 67 | KIF15 | 610731 4070731 6520014 | 5200 | 0.064 | 0.4254 | No | ||
| 68 | NBN | 730538 2470619 4780594 | 5294 | 0.060 | 0.4211 | No | ||
| 69 | EPGN | 6980450 | 5333 | 0.059 | 0.4196 | No | ||
| 70 | TTK | 3800129 | 6467 | 0.026 | 0.3586 | No | ||
| 71 | CENPF | 5050088 | 6665 | 0.022 | 0.3482 | No | ||
| 72 | PAM | 5290528 | 6751 | 0.020 | 0.3438 | No | ||
| 73 | ANAPC10 | 870086 1170037 2260129 | 6773 | 0.019 | 0.3429 | No | ||
| 74 | RAN | 2260446 4590647 | 6789 | 0.019 | 0.3423 | No | ||
| 75 | CDKN1A | 4050088 6400706 | 6815 | 0.019 | 0.3411 | No | ||
| 76 | KIF2C | 6940082 | 6866 | 0.018 | 0.3386 | No | ||
| 77 | BTG3 | 7050079 | 7663 | 0.004 | 0.2956 | No | ||
| 78 | EREG | 50519 4920129 | 7933 | -0.001 | 0.2811 | No | ||
| 79 | BUB1 | 5390270 | 8457 | -0.009 | 0.2529 | No | ||
| 80 | POLA1 | 4760541 | 9007 | -0.018 | 0.2234 | No | ||
| 81 | FOXC1 | 580239 | 9302 | -0.024 | 0.2077 | No | ||
| 82 | KIF11 | 5390139 | 9371 | -0.025 | 0.2043 | No | ||
| 83 | CDKN3 | 110520 | 10134 | -0.039 | 0.1635 | No | ||
| 84 | TTN | 2320161 4670056 6550026 | 10145 | -0.039 | 0.1634 | No | ||
| 85 | PRMT5 | 2760180 4590072 | 10509 | -0.047 | 0.1443 | No | ||
| 86 | SNF1LK | 6110403 | 10563 | -0.048 | 0.1419 | No | ||
| 87 | CETN1 | 6020056 | 10660 | -0.050 | 0.1372 | No | ||
| 88 | APBB2 | 5130403 | 10683 | -0.051 | 0.1366 | No | ||
| 89 | KATNA1 | 6290048 | 10704 | -0.051 | 0.1361 | No | ||
| 90 | NEK2 | 2120520 | 10893 | -0.056 | 0.1265 | No | ||
| 91 | PAFAH1B1 | 4230333 6420121 6450066 | 11397 | -0.070 | 0.1000 | No | ||
| 92 | CLIP1 | 2850162 6770397 | 11827 | -0.084 | 0.0777 | No | ||
| 93 | CUL5 | 450142 | 12089 | -0.094 | 0.0646 | No | ||
| 94 | NEK6 | 3360687 | 12419 | -0.109 | 0.0480 | No | ||
| 95 | SPHK1 | 2470113 4230398 6220397 | 12538 | -0.115 | 0.0428 | No | ||
| 96 | CENPE | 2850022 | 12747 | -0.126 | 0.0329 | No | ||
| 97 | TBRG4 | 3610239 6370594 | 12866 | -0.133 | 0.0280 | No | ||
| 98 | CDCA5 | 5670131 | 13200 | -0.156 | 0.0116 | No | ||
| 99 | PLK1 | 1780369 2640121 | 13227 | -0.159 | 0.0119 | No | ||
| 100 | UBE2C | 6130017 | 13412 | -0.173 | 0.0038 | No | ||
| 101 | ANLN | 1780113 2470537 6840047 | 13567 | -0.187 | -0.0025 | No | ||
| 102 | CDKN2B | 6020040 | 13572 | -0.187 | -0.0007 | No | ||
| 103 | ANAPC11 | 1780601 | 13588 | -0.189 | 0.0005 | No | ||
| 104 | DLG1 | 2630091 6020286 | 13610 | -0.191 | 0.0014 | No | ||
| 105 | ACVR1 | 6840671 | 13964 | -0.228 | -0.0153 | No | ||
| 106 | RCC1 | 2480358 | 14128 | -0.246 | -0.0214 | No | ||
| 107 | ASNS | 110368 7100687 | 14169 | -0.253 | -0.0209 | No | ||
| 108 | TPD52L1 | 4590711 5570575 | 14225 | -0.260 | -0.0211 | No | ||
| 109 | CUL2 | 4200278 | 14495 | -0.298 | -0.0325 | No | ||
| 110 | DCTN2 | 540471 3780717 | 14688 | -0.333 | -0.0393 | No | ||
| 111 | CDKN2D | 6040035 | 14900 | -0.370 | -0.0467 | No | ||
| 112 | APBB1 | 2690338 | 15141 | -0.422 | -0.0552 | No | ||
| 113 | CDK6 | 4920253 | 15335 | -0.469 | -0.0606 | No | ||
| 114 | E2F1 | 5360093 | 15379 | -0.483 | -0.0578 | No | ||
| 115 | NUMA1 | 6520576 | 15578 | -0.548 | -0.0626 | No | ||
| 116 | TGFB1 | 1940162 | 15749 | -0.610 | -0.0653 | No | ||
| 117 | CDKN2C | 5050750 5130148 | 15989 | -0.707 | -0.0707 | No | ||
| 118 | TARDBP | 870390 2350093 | 16452 | -0.917 | -0.0859 | No | ||
| 119 | POLD1 | 4830026 | 16821 | -1.079 | -0.0942 | No | ||
| 120 | MAP3K11 | 7000039 | 17023 | -1.210 | -0.0922 | No | ||
| 121 | CHFR | 3390050 6450300 6450736 | 17127 | -1.272 | -0.0841 | No | ||
| 122 | CDC25B | 6940102 | 17519 | -1.538 | -0.0888 | No | ||
| 123 | CHMP1A | 5550441 | 17651 | -1.647 | -0.0783 | No | ||
| 124 | MAD2L2 | 1240358 | 17788 | -1.784 | -0.0666 | No | ||
| 125 | CDK4 | 540075 4540600 | 18101 | -2.120 | -0.0608 | No | ||
| 126 | SMAD3 | 6450671 | 18195 | -2.238 | -0.0419 | No | ||
| 127 | BCAT1 | 3290128 4050408 | 18470 | -2.890 | -0.0258 | No | ||
| 128 | CDK10 | 60164 1190717 3130128 3990044 | 18523 | -3.142 | 0.0050 | No |

