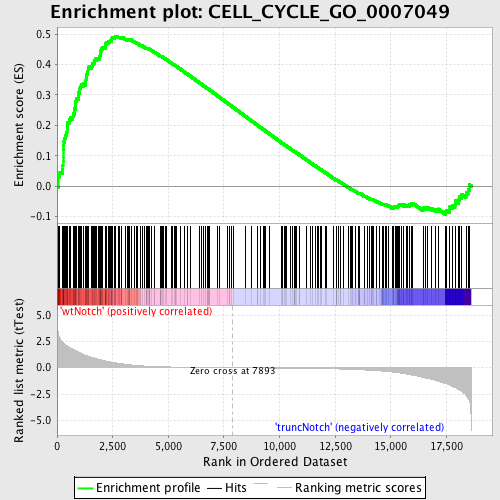
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | CELL_CYCLE_GO_0007049 |
| Enrichment Score (ES) | 0.49356252 |
| Normalized Enrichment Score (NES) | 1.6201555 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.28862822 |
| FWER p-Value | 0.944 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NCAPH | 6220435 | 56 | 3.183 | 0.0153 | Yes | ||
| 2 | AKAP8 | 4210026 | 57 | 3.171 | 0.0337 | Yes | ||
| 3 | KPNA2 | 3450114 | 125 | 2.781 | 0.0461 | Yes | ||
| 4 | RABGAP1 | 5050397 | 236 | 2.466 | 0.0543 | Yes | ||
| 5 | BUB1B | 1450288 | 261 | 2.413 | 0.0669 | Yes | ||
| 6 | POLE | 6020538 | 265 | 2.398 | 0.0806 | Yes | ||
| 7 | GTPBP4 | 6840332 | 269 | 2.393 | 0.0943 | Yes | ||
| 8 | ANAPC4 | 4540338 | 272 | 2.384 | 0.1080 | Yes | ||
| 9 | MAD2L1 | 4480725 | 273 | 2.379 | 0.1217 | Yes | ||
| 10 | NASP | 2260139 2940369 5130707 | 298 | 2.326 | 0.1338 | Yes | ||
| 11 | TPX2 | 6420324 | 306 | 2.306 | 0.1468 | Yes | ||
| 12 | CCND1 | 460524 770309 3120576 6980398 | 346 | 2.224 | 0.1575 | Yes | ||
| 13 | RB1 | 5900338 | 390 | 2.152 | 0.1676 | Yes | ||
| 14 | POLS | 5890438 | 424 | 2.095 | 0.1779 | Yes | ||
| 15 | CUL3 | 1850520 | 446 | 2.066 | 0.1887 | Yes | ||
| 16 | HCFC1 | 2260008 | 463 | 2.041 | 0.1996 | Yes | ||
| 17 | SKP2 | 360711 380093 4810368 | 483 | 2.012 | 0.2102 | Yes | ||
| 18 | SESN1 | 2120021 | 536 | 1.926 | 0.2185 | Yes | ||
| 19 | ANAPC5 | 730164 7100685 | 597 | 1.862 | 0.2260 | Yes | ||
| 20 | NUSAP1 | 940048 3120435 | 714 | 1.733 | 0.2296 | Yes | ||
| 21 | CDC7 | 4060546 4850041 | 725 | 1.718 | 0.2390 | Yes | ||
| 22 | CUL1 | 1990632 | 772 | 1.677 | 0.2462 | Yes | ||
| 23 | PTPRC | 130402 5290148 | 788 | 1.663 | 0.2550 | Yes | ||
| 24 | MNAT1 | 730292 2350364 | 831 | 1.616 | 0.2620 | Yes | ||
| 25 | PPP6C | 2450670 | 840 | 1.613 | 0.2709 | Yes | ||
| 26 | ATR | 6860273 | 844 | 1.606 | 0.2800 | Yes | ||
| 27 | NDC80 | 4120465 | 867 | 1.583 | 0.2880 | Yes | ||
| 28 | CETN3 | 3170402 | 948 | 1.485 | 0.2922 | Yes | ||
| 29 | ZWINT | 6940670 | 954 | 1.481 | 0.3005 | Yes | ||
| 30 | PRC1 | 870092 5890204 | 962 | 1.472 | 0.3086 | Yes | ||
| 31 | KIF23 | 5570112 | 1019 | 1.409 | 0.3137 | Yes | ||
| 32 | GSPT1 | 5420050 | 1023 | 1.408 | 0.3217 | Yes | ||
| 33 | CCNE2 | 3120537 | 1049 | 1.388 | 0.3283 | Yes | ||
| 34 | CDKN1C | 6520577 | 1081 | 1.353 | 0.3344 | Yes | ||
| 35 | MPHOSPH6 | 5570164 | 1179 | 1.259 | 0.3364 | Yes | ||
| 36 | LIG3 | 2940300 | 1266 | 1.187 | 0.3386 | Yes | ||
| 37 | CDT1 | 3780682 | 1268 | 1.186 | 0.3454 | Yes | ||
| 38 | KHDRBS1 | 1240403 6040040 | 1297 | 1.152 | 0.3505 | Yes | ||
| 39 | HSPA2 | 6420592 | 1304 | 1.149 | 0.3568 | Yes | ||
| 40 | AURKA | 780537 | 1317 | 1.140 | 0.3628 | Yes | ||
| 41 | SMC1A | 3060600 5700148 5890113 6370154 | 1333 | 1.130 | 0.3685 | Yes | ||
| 42 | ZW10 | 2900735 3520687 | 1362 | 1.103 | 0.3733 | Yes | ||
| 43 | CDKN2A | 4670215 4760047 | 1380 | 1.090 | 0.3787 | Yes | ||
| 44 | USP16 | 2030671 | 1388 | 1.082 | 0.3846 | Yes | ||
| 45 | PTEN | 3390064 | 1409 | 1.066 | 0.3896 | Yes | ||
| 46 | TIPIN | 1570301 2480113 | 1432 | 1.048 | 0.3945 | Yes | ||
| 47 | PCBP4 | 6350722 | 1563 | 0.991 | 0.3931 | Yes | ||
| 48 | CD28 | 1400739 4210093 | 1578 | 0.983 | 0.3980 | Yes | ||
| 49 | CDC16 | 1940706 | 1589 | 0.980 | 0.4031 | Yes | ||
| 50 | KNTC1 | 430079 | 1652 | 0.940 | 0.4052 | Yes | ||
| 51 | EIF4G2 | 3800575 6860184 | 1689 | 0.918 | 0.4085 | Yes | ||
| 52 | GMNN | 2630148 | 1691 | 0.916 | 0.4138 | Yes | ||
| 53 | RAD51L3 | 2230609 2810286 3190008 3390053 4230168 | 1703 | 0.906 | 0.4184 | Yes | ||
| 54 | RAD21 | 1990278 | 1781 | 0.856 | 0.4192 | Yes | ||
| 55 | PCAF | 2230161 2570369 6550451 | 1851 | 0.812 | 0.4201 | Yes | ||
| 56 | BIRC5 | 110408 580014 1770632 | 1894 | 0.790 | 0.4224 | Yes | ||
| 57 | ABL1 | 1050593 2030050 4010114 | 1898 | 0.789 | 0.4268 | Yes | ||
| 58 | CDK2AP1 | 2340156 | 1928 | 0.770 | 0.4296 | Yes | ||
| 59 | DLG7 | 3120041 | 1942 | 0.763 | 0.4333 | Yes | ||
| 60 | CDK5R1 | 3870161 | 1949 | 0.758 | 0.4374 | Yes | ||
| 61 | MRE11A | 3850601 4670332 | 1958 | 0.748 | 0.4413 | Yes | ||
| 62 | CDC37 | 2570537 | 1969 | 0.742 | 0.4450 | Yes | ||
| 63 | CCNG2 | 3190095 | 1989 | 0.734 | 0.4482 | Yes | ||
| 64 | TBRG1 | 6110678 6520181 | 1993 | 0.729 | 0.4523 | Yes | ||
| 65 | NDE1 | 1580242 3830097 | 2027 | 0.703 | 0.4545 | Yes | ||
| 66 | CDC45L | 70537 3130114 | 2050 | 0.690 | 0.4573 | Yes | ||
| 67 | CDC6 | 4570296 5360600 | 2159 | 0.641 | 0.4551 | Yes | ||
| 68 | MADD | 6100725 7000088 | 2162 | 0.638 | 0.4587 | Yes | ||
| 69 | CCND3 | 380528 730500 | 2164 | 0.637 | 0.4623 | Yes | ||
| 70 | STMN1 | 1990717 | 2167 | 0.636 | 0.4659 | Yes | ||
| 71 | CCNA2 | 5290075 | 2176 | 0.634 | 0.4691 | Yes | ||
| 72 | CDC25A | 3800184 | 2212 | 0.617 | 0.4708 | Yes | ||
| 73 | MYH10 | 2640673 | 2217 | 0.614 | 0.4741 | Yes | ||
| 74 | RAD50 | 4050184 6770746 | 2314 | 0.577 | 0.4722 | Yes | ||
| 75 | RASSF1 | 2760364 4850725 | 2324 | 0.574 | 0.4750 | Yes | ||
| 76 | ING4 | 510632 2340619 2850176 5290706 | 2368 | 0.554 | 0.4759 | Yes | ||
| 77 | ERCC3 | 6900008 | 2382 | 0.551 | 0.4784 | Yes | ||
| 78 | HUS1 | 3440494 3800156 | 2429 | 0.533 | 0.4789 | Yes | ||
| 79 | SUGT1 | 1690670 | 2430 | 0.532 | 0.4820 | Yes | ||
| 80 | CIT | 2370601 | 2435 | 0.528 | 0.4848 | Yes | ||
| 81 | SMC4 | 5910240 | 2448 | 0.524 | 0.4872 | Yes | ||
| 82 | EGF | 5220154 | 2474 | 0.515 | 0.4888 | Yes | ||
| 83 | CUL4A | 1170088 2320008 2470278 | 2506 | 0.504 | 0.4900 | Yes | ||
| 84 | APC | 3850484 5860722 | 2589 | 0.475 | 0.4883 | Yes | ||
| 85 | STRN3 | 1450093 6200685 | 2598 | 0.471 | 0.4906 | Yes | ||
| 86 | SSSCA1 | 1240402 780309 | 2608 | 0.469 | 0.4928 | Yes | ||
| 87 | XRCC2 | 4920706 | 2644 | 0.458 | 0.4936 | Yes | ||
| 88 | GTF2H1 | 2570746 2650148 | 2751 | 0.423 | 0.4902 | No | ||
| 89 | PPP5C | 3130047 | 2795 | 0.409 | 0.4902 | No | ||
| 90 | NOLC1 | 2350195 | 2875 | 0.381 | 0.4881 | No | ||
| 91 | GFI1 | 730180 | 2907 | 0.371 | 0.4886 | No | ||
| 92 | AIF1 | 130332 5220017 | 2913 | 0.369 | 0.4905 | No | ||
| 93 | KIF22 | 1190368 | 3067 | 0.321 | 0.4840 | No | ||
| 94 | LATS2 | 4480593 6020494 | 3166 | 0.294 | 0.4803 | No | ||
| 95 | RAD17 | 5220739 | 3194 | 0.288 | 0.4805 | No | ||
| 96 | CDK2 | 130484 2260301 4010088 5050110 | 3216 | 0.284 | 0.4810 | No | ||
| 97 | TUBG1 | 3140204 | 3242 | 0.280 | 0.4813 | No | ||
| 98 | BRCA2 | 4280372 | 3259 | 0.274 | 0.4820 | No | ||
| 99 | ACVR1B | 3610446 5570195 | 3271 | 0.272 | 0.4830 | No | ||
| 100 | UBB | 4810138 | 3327 | 0.260 | 0.4815 | No | ||
| 101 | RAD51 | 6110450 6980280 | 3499 | 0.225 | 0.4734 | No | ||
| 102 | RAD51L1 | 4010112 5860435 | 3548 | 0.213 | 0.4721 | No | ||
| 103 | GFI1B | 4570131 | 3607 | 0.204 | 0.4701 | No | ||
| 104 | GAS7 | 2120358 6450494 | 3737 | 0.183 | 0.4641 | No | ||
| 105 | PIN1 | 2970408 | 3758 | 0.180 | 0.4640 | No | ||
| 106 | TGFA | 1570332 | 3820 | 0.172 | 0.4617 | No | ||
| 107 | SMC3 | 870546 1400411 5700039 6450577 | 3910 | 0.158 | 0.4578 | No | ||
| 108 | NPM2 | 6040139 | 4003 | 0.148 | 0.4536 | No | ||
| 109 | MAP2K6 | 1230056 2940204 | 4023 | 0.145 | 0.4534 | No | ||
| 110 | ERN1 | 2360403 | 4031 | 0.143 | 0.4539 | No | ||
| 111 | CDKN1B | 3800025 6450044 | 4044 | 0.142 | 0.4540 | No | ||
| 112 | CCNG1 | 870402 1500685 6110594 | 4108 | 0.135 | 0.4514 | No | ||
| 113 | DDX11 | 6590671 | 4135 | 0.133 | 0.4507 | No | ||
| 114 | NOTCH2 | 2570397 | 4149 | 0.131 | 0.4508 | No | ||
| 115 | SH3BP4 | 3140242 | 4155 | 0.131 | 0.4513 | No | ||
| 116 | CDC25C | 2570673 4760161 6520707 | 4241 | 0.124 | 0.4474 | No | ||
| 117 | CCNT2 | 2100390 4150563 | 4381 | 0.111 | 0.4404 | No | ||
| 118 | USH1C | 1570372 4230364 | 4394 | 0.110 | 0.4404 | No | ||
| 119 | PML | 50093 2190435 2450402 3840082 | 4633 | 0.093 | 0.4280 | No | ||
| 120 | FBXO5 | 2630551 | 4653 | 0.092 | 0.4275 | No | ||
| 121 | TUBE1 | 2360079 6520685 | 4664 | 0.091 | 0.4275 | No | ||
| 122 | ATM | 3610110 4050524 | 4686 | 0.090 | 0.4268 | No | ||
| 123 | UPF1 | 430164 1190750 | 4728 | 0.087 | 0.4251 | No | ||
| 124 | DCTN3 | 6840131 | 4767 | 0.085 | 0.4235 | No | ||
| 125 | SYCP1 | 2570010 | 4855 | 0.080 | 0.4192 | No | ||
| 126 | PIM2 | 840136 5720692 7100577 | 4931 | 0.076 | 0.4156 | No | ||
| 127 | CDC23 | 3190593 | 5157 | 0.066 | 0.4037 | No | ||
| 128 | KIF15 | 610731 4070731 6520014 | 5200 | 0.064 | 0.4018 | No | ||
| 129 | NBN | 730538 2470619 4780594 | 5294 | 0.060 | 0.3971 | No | ||
| 130 | CHEK1 | 50167 1940300 | 5327 | 0.059 | 0.3957 | No | ||
| 131 | EPGN | 6980450 | 5333 | 0.059 | 0.3957 | No | ||
| 132 | CCNK | 6980048 | 5366 | 0.057 | 0.3943 | No | ||
| 133 | UHRF2 | 60050 840280 1340746 5080091 | 5533 | 0.051 | 0.3856 | No | ||
| 134 | GPS2 | 5910672 | 5720 | 0.045 | 0.3757 | No | ||
| 135 | RAD52 | 110093 | 5862 | 0.041 | 0.3683 | No | ||
| 136 | GAS1 | 2120504 | 5869 | 0.041 | 0.3682 | No | ||
| 137 | BMP4 | 380113 | 6000 | 0.037 | 0.3613 | No | ||
| 138 | MYC | 380541 4670170 | 6382 | 0.028 | 0.3407 | No | ||
| 139 | TTK | 3800129 | 6467 | 0.026 | 0.3363 | No | ||
| 140 | SPO11 | 540288 4780239 | 6593 | 0.023 | 0.3296 | No | ||
| 141 | CENPF | 5050088 | 6665 | 0.022 | 0.3258 | No | ||
| 142 | PAM | 5290528 | 6751 | 0.020 | 0.3213 | No | ||
| 143 | ANAPC10 | 870086 1170037 2260129 | 6773 | 0.019 | 0.3203 | No | ||
| 144 | RAN | 2260446 4590647 | 6789 | 0.019 | 0.3196 | No | ||
| 145 | CDKN1A | 4050088 6400706 | 6815 | 0.019 | 0.3183 | No | ||
| 146 | RAD9A | 110300 1940632 3990390 6040014 | 6846 | 0.018 | 0.3168 | No | ||
| 147 | KIF2C | 6940082 | 6866 | 0.018 | 0.3159 | No | ||
| 148 | MLF1 | 4570722 | 7190 | 0.012 | 0.2983 | No | ||
| 149 | MDM4 | 4070504 4780008 | 7308 | 0.010 | 0.2920 | No | ||
| 150 | BTG3 | 7050079 | 7663 | 0.004 | 0.2727 | No | ||
| 151 | JMY | 3360242 | 7741 | 0.003 | 0.2686 | No | ||
| 152 | DHRS2 | 4200577 | 7750 | 0.002 | 0.2681 | No | ||
| 153 | CCNA1 | 6290113 | 7823 | 0.001 | 0.2642 | No | ||
| 154 | EREG | 50519 4920129 | 7933 | -0.001 | 0.2583 | No | ||
| 155 | BUB1 | 5390270 | 8457 | -0.009 | 0.2298 | No | ||
| 156 | CCND2 | 5340167 | 8718 | -0.014 | 0.2158 | No | ||
| 157 | MSH4 | 3940068 | 8994 | -0.018 | 0.2009 | No | ||
| 158 | POLA1 | 4760541 | 9007 | -0.018 | 0.2003 | No | ||
| 159 | NPM1 | 4730427 | 9124 | -0.020 | 0.1941 | No | ||
| 160 | EGFL6 | 3940181 | 9267 | -0.023 | 0.1865 | No | ||
| 161 | FOXC1 | 580239 | 9302 | -0.024 | 0.1848 | No | ||
| 162 | KIF11 | 5390139 | 9371 | -0.025 | 0.1812 | No | ||
| 163 | CKS2 | 1410156 | 9553 | -0.028 | 0.1715 | No | ||
| 164 | TIMELESS | 3710315 | 10080 | -0.038 | 0.1431 | No | ||
| 165 | CDKN3 | 110520 | 10134 | -0.039 | 0.1404 | No | ||
| 166 | TTN | 2320161 4670056 6550026 | 10145 | -0.039 | 0.1401 | No | ||
| 167 | ALOX15B | 450736 | 10218 | -0.040 | 0.1364 | No | ||
| 168 | INHA | 6100102 | 10284 | -0.042 | 0.1331 | No | ||
| 169 | BMP2 | 6620687 | 10322 | -0.042 | 0.1313 | No | ||
| 170 | PRMT5 | 2760180 4590072 | 10509 | -0.047 | 0.1215 | No | ||
| 171 | SNF1LK | 6110403 | 10563 | -0.048 | 0.1189 | No | ||
| 172 | CETN1 | 6020056 | 10660 | -0.050 | 0.1139 | No | ||
| 173 | PPP1R9B | 3130619 | 10678 | -0.051 | 0.1133 | No | ||
| 174 | APBB2 | 5130403 | 10683 | -0.051 | 0.1134 | No | ||
| 175 | KATNA1 | 6290048 | 10704 | -0.051 | 0.1126 | No | ||
| 176 | RPRM | 1580706 | 10761 | -0.053 | 0.1098 | No | ||
| 177 | CDK7 | 2640451 | 10877 | -0.056 | 0.1039 | No | ||
| 178 | NEK2 | 2120520 | 10893 | -0.056 | 0.1034 | No | ||
| 179 | HERC5 | 4280687 | 11223 | -0.065 | 0.0858 | No | ||
| 180 | PAFAH1B1 | 4230333 6420121 6450066 | 11397 | -0.070 | 0.0768 | No | ||
| 181 | CDK5R2 | 3800110 | 11457 | -0.072 | 0.0740 | No | ||
| 182 | GPS1 | 2470037 4760315 6200528 7100040 | 11635 | -0.077 | 0.0648 | No | ||
| 183 | XPC | 6370193 | 11703 | -0.079 | 0.0616 | No | ||
| 184 | BTG4 | 4060020 | 11764 | -0.082 | 0.0588 | No | ||
| 185 | CLIP1 | 2850162 6770397 | 11827 | -0.084 | 0.0559 | No | ||
| 186 | TGFB2 | 4920292 | 11864 | -0.085 | 0.0545 | No | ||
| 187 | DMC1 | 450341 | 12054 | -0.092 | 0.0447 | No | ||
| 188 | CUL5 | 450142 | 12089 | -0.094 | 0.0434 | No | ||
| 189 | NEK6 | 3360687 | 12419 | -0.109 | 0.0261 | No | ||
| 190 | SPHK1 | 2470113 4230398 6220397 | 12538 | -0.115 | 0.0203 | No | ||
| 191 | ANAPC2 | 6660438 | 12550 | -0.116 | 0.0204 | No | ||
| 192 | PPP1R13B | 4010605 4760181 | 12552 | -0.116 | 0.0210 | No | ||
| 193 | RUNX3 | 3800546 | 12636 | -0.121 | 0.0172 | No | ||
| 194 | RAD1 | 4200551 | 12723 | -0.125 | 0.0132 | No | ||
| 195 | CENPE | 2850022 | 12747 | -0.126 | 0.0127 | No | ||
| 196 | TBRG4 | 3610239 6370594 | 12866 | -0.133 | 0.0070 | No | ||
| 197 | CCNT1 | 2450563 5390184 | 13093 | -0.149 | -0.0044 | No | ||
| 198 | CDCA5 | 5670131 | 13200 | -0.156 | -0.0093 | No | ||
| 199 | PLK1 | 1780369 2640121 | 13227 | -0.159 | -0.0098 | No | ||
| 200 | FANCG | 4010471 | 13265 | -0.161 | -0.0109 | No | ||
| 201 | UBE2C | 6130017 | 13412 | -0.173 | -0.0178 | No | ||
| 202 | AHR | 4560671 | 13546 | -0.185 | -0.0240 | No | ||
| 203 | ANLN | 1780113 2470537 6840047 | 13567 | -0.187 | -0.0240 | No | ||
| 204 | CDKN2B | 6020040 | 13572 | -0.187 | -0.0232 | No | ||
| 205 | ANAPC11 | 1780601 | 13588 | -0.189 | -0.0229 | No | ||
| 206 | DLG1 | 2630091 6020286 | 13610 | -0.191 | -0.0229 | No | ||
| 207 | DUSP13 | 6400152 | 13809 | -0.211 | -0.0325 | No | ||
| 208 | CITED2 | 5670114 5130088 | 13832 | -0.213 | -0.0325 | No | ||
| 209 | ACVR1 | 6840671 | 13964 | -0.228 | -0.0383 | No | ||
| 210 | PPM1G | 610725 | 14046 | -0.236 | -0.0414 | No | ||
| 211 | PLAGL1 | 3190082 6200193 | 14114 | -0.243 | -0.0436 | No | ||
| 212 | RCC1 | 2480358 | 14128 | -0.246 | -0.0429 | No | ||
| 213 | ASNS | 110368 7100687 | 14169 | -0.253 | -0.0436 | No | ||
| 214 | TPD52L1 | 4590711 5570575 | 14225 | -0.260 | -0.0451 | No | ||
| 215 | RAD54L | 2970041 | 14358 | -0.276 | -0.0507 | No | ||
| 216 | NEK11 | 5360369 7380521 | 14373 | -0.278 | -0.0499 | No | ||
| 217 | CUL2 | 4200278 | 14495 | -0.298 | -0.0547 | No | ||
| 218 | MAPK12 | 450022 1340717 7050484 | 14604 | -0.318 | -0.0588 | No | ||
| 219 | UHMK1 | 3850670 6450064 | 14639 | -0.324 | -0.0588 | No | ||
| 220 | DCTN2 | 540471 3780717 | 14688 | -0.333 | -0.0595 | No | ||
| 221 | TBX3 | 2570672 | 14782 | -0.348 | -0.0625 | No | ||
| 222 | PFDN1 | 7000095 | 14801 | -0.350 | -0.0615 | No | ||
| 223 | CDKN2D | 6040035 | 14900 | -0.370 | -0.0647 | No | ||
| 224 | SERTAD1 | 2680332 | 15068 | -0.408 | -0.0714 | No | ||
| 225 | CDC20 | 3440017 3440044 6220088 | 15103 | -0.413 | -0.0709 | No | ||
| 226 | CENTD2 | 60408 2510156 6100494 | 15112 | -0.415 | -0.0689 | No | ||
| 227 | APBB1 | 2690338 | 15141 | -0.422 | -0.0680 | No | ||
| 228 | RACGAP1 | 3990162 6620736 | 15208 | -0.437 | -0.0691 | No | ||
| 229 | TP53 | 6130707 | 15236 | -0.442 | -0.0680 | No | ||
| 230 | MCM2 | 5050139 | 15297 | -0.459 | -0.0686 | No | ||
| 231 | CHEK2 | 610139 1050022 | 15319 | -0.465 | -0.0671 | No | ||
| 232 | CDK6 | 4920253 | 15335 | -0.469 | -0.0652 | No | ||
| 233 | TOP3A | 7000435 | 15350 | -0.476 | -0.0632 | No | ||
| 234 | E2F1 | 5360093 | 15379 | -0.483 | -0.0619 | No | ||
| 235 | CDK5RAP1 | 2570471 4010465 5960161 | 15406 | -0.491 | -0.0605 | No | ||
| 236 | GADD45A | 2900717 | 15472 | -0.512 | -0.0611 | No | ||
| 237 | BOLL | 6100743 | 15568 | -0.545 | -0.0631 | No | ||
| 238 | NUMA1 | 6520576 | 15578 | -0.548 | -0.0604 | No | ||
| 239 | HPGD | 6770192 | 15714 | -0.599 | -0.0643 | No | ||
| 240 | TGFB1 | 1940162 | 15749 | -0.610 | -0.0627 | No | ||
| 241 | PA2G4 | 2030463 | 15839 | -0.638 | -0.0638 | No | ||
| 242 | CDK5RAP3 | 3130576 | 15852 | -0.643 | -0.0608 | No | ||
| 243 | RHOB | 1500309 | 15922 | -0.678 | -0.0606 | No | ||
| 244 | STAG3 | 3830112 | 15933 | -0.682 | -0.0572 | No | ||
| 245 | CDKN2C | 5050750 5130148 | 15989 | -0.707 | -0.0561 | No | ||
| 246 | TARDBP | 870390 2350093 | 16452 | -0.917 | -0.0760 | No | ||
| 247 | DDB1 | 6110687 | 16471 | -0.927 | -0.0716 | No | ||
| 248 | STK11 | 6550278 | 16569 | -0.974 | -0.0713 | No | ||
| 249 | MFN2 | 2260195 6100164 | 16642 | -1.000 | -0.0694 | No | ||
| 250 | POLD1 | 4830026 | 16821 | -1.079 | -0.0729 | No | ||
| 251 | MAP3K11 | 7000039 | 17023 | -1.210 | -0.0769 | No | ||
| 252 | CHFR | 3390050 6450300 6450736 | 17127 | -1.272 | -0.0751 | No | ||
| 253 | BCCIP | 670450 2650750 5270519 6660711 | 17460 | -1.494 | -0.0846 | No | ||
| 254 | CDC25B | 6940102 | 17519 | -1.538 | -0.0789 | No | ||
| 255 | CHMP1A | 5550441 | 17651 | -1.647 | -0.0765 | No | ||
| 256 | MSH5 | 1340019 4730333 | 17655 | -1.651 | -0.0671 | No | ||
| 257 | MAD2L2 | 1240358 | 17788 | -1.784 | -0.0640 | No | ||
| 258 | ZBTB17 | 6220082 | 17885 | -1.873 | -0.0584 | No | ||
| 259 | ERCC2 | 2360750 4060390 6550138 | 17904 | -1.897 | -0.0484 | No | ||
| 260 | BMP7 | 870039 | 18045 | -2.053 | -0.0442 | No | ||
| 261 | CDK4 | 540075 4540600 | 18101 | -2.120 | -0.0350 | No | ||
| 262 | SMAD3 | 6450671 | 18195 | -2.238 | -0.0271 | No | ||
| 263 | DTYMK | 2340377 | 18381 | -2.603 | -0.0221 | No | ||
| 264 | BCAT1 | 3290128 4050408 | 18470 | -2.890 | -0.0102 | No | ||
| 265 | CDK10 | 60164 1190717 3130128 3990044 | 18523 | -3.142 | 0.0051 | No |

