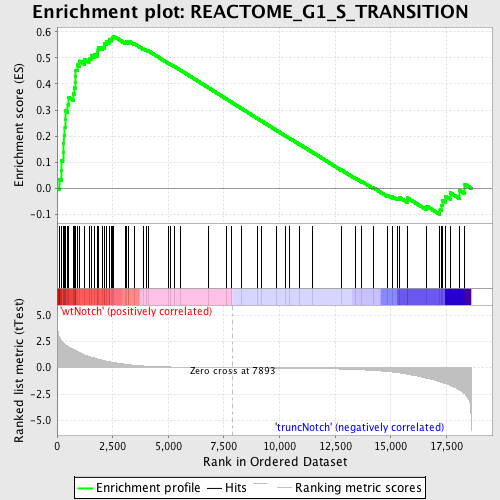
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | REACTOME_G1_S_TRANSITION |
| Enrichment Score (ES) | 0.5835186 |
| Normalized Enrichment Score (NES) | 1.6513255 |
| Nominal p-value | 0.0026809652 |
| FDR q-value | 0.18116543 |
| FWER p-Value | 0.781 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMD14 | 5690593 | 119 | 2.806 | 0.0352 | Yes | ||
| 2 | PSMA4 | 4560427 | 198 | 2.539 | 0.0686 | Yes | ||
| 3 | CCNB1 | 4590433 4780372 | 211 | 2.518 | 0.1052 | Yes | ||
| 4 | POLE | 6020538 | 265 | 2.398 | 0.1379 | Yes | ||
| 5 | PSMD7 | 2030619 6220594 | 293 | 2.337 | 0.1711 | Yes | ||
| 6 | CCNH | 3190347 | 309 | 2.289 | 0.2042 | Yes | ||
| 7 | PSMA2 | 6510093 | 353 | 2.215 | 0.2347 | Yes | ||
| 8 | RPA1 | 360452 | 385 | 2.159 | 0.2650 | Yes | ||
| 9 | RB1 | 5900338 | 390 | 2.152 | 0.2967 | Yes | ||
| 10 | PSMD12 | 730044 | 488 | 2.007 | 0.3212 | Yes | ||
| 11 | PCNA | 940754 | 526 | 1.941 | 0.3480 | Yes | ||
| 12 | CDC7 | 4060546 4850041 | 725 | 1.718 | 0.3627 | Yes | ||
| 13 | CUL1 | 1990632 | 772 | 1.677 | 0.3851 | Yes | ||
| 14 | RPA3 | 5700136 | 821 | 1.631 | 0.4067 | Yes | ||
| 15 | MNAT1 | 730292 2350364 | 831 | 1.616 | 0.4301 | Yes | ||
| 16 | PSMD10 | 520494 1170576 3830050 | 842 | 1.611 | 0.4535 | Yes | ||
| 17 | MCM4 | 2760673 5420711 | 904 | 1.538 | 0.4730 | Yes | ||
| 18 | ORC5L | 1940133 1940711 | 1013 | 1.414 | 0.4881 | Yes | ||
| 19 | PSMD4 | 430068 | 1236 | 1.214 | 0.4941 | Yes | ||
| 20 | RRM2 | 6350059 6940162 | 1434 | 1.046 | 0.4990 | Yes | ||
| 21 | PRIM1 | 6420746 | 1539 | 0.999 | 0.5082 | Yes | ||
| 22 | TFDP1 | 1980112 | 1671 | 0.926 | 0.5149 | Yes | ||
| 23 | MCM6 | 60092 540181 6510110 | 1822 | 0.831 | 0.5191 | Yes | ||
| 24 | PSME1 | 450193 4480035 | 1836 | 0.823 | 0.5306 | Yes | ||
| 25 | PSMD3 | 1400647 | 1854 | 0.811 | 0.5417 | Yes | ||
| 26 | CDC45L | 70537 3130114 | 2050 | 0.690 | 0.5414 | Yes | ||
| 27 | E2F2 | 5270609 5570377 7000465 | 2122 | 0.660 | 0.5473 | Yes | ||
| 28 | PSMC5 | 2760315 6550021 | 2143 | 0.650 | 0.5559 | Yes | ||
| 29 | CDC25A | 3800184 | 2212 | 0.617 | 0.5613 | Yes | ||
| 30 | MCM7 | 3290292 5220056 | 2334 | 0.569 | 0.5632 | Yes | ||
| 31 | PSMC6 | 2810021 | 2356 | 0.560 | 0.5704 | Yes | ||
| 32 | PSMC4 | 580050 1580025 | 2463 | 0.518 | 0.5723 | Yes | ||
| 33 | PSME3 | 2810537 | 2475 | 0.515 | 0.5794 | Yes | ||
| 34 | ORC4L | 4230538 5550288 | 2535 | 0.494 | 0.5835 | Yes | ||
| 35 | PSMB1 | 2940402 | 3086 | 0.317 | 0.5586 | No | ||
| 36 | PSMB2 | 940035 4210324 | 3097 | 0.313 | 0.5627 | No | ||
| 37 | CDK2 | 130484 2260301 4010088 5050110 | 3216 | 0.284 | 0.5605 | No | ||
| 38 | RPA2 | 2760301 5420195 | 3221 | 0.283 | 0.5645 | No | ||
| 39 | ORC1L | 2370328 6110390 | 3473 | 0.230 | 0.5543 | No | ||
| 40 | WEE1 | 3390070 | 3884 | 0.160 | 0.5346 | No | ||
| 41 | ORC2L | 1990470 6510019 | 4029 | 0.144 | 0.5290 | No | ||
| 42 | PSMC2 | 7040010 | 4109 | 0.135 | 0.5267 | No | ||
| 43 | PSMD11 | 2340538 6510053 | 5028 | 0.072 | 0.4783 | No | ||
| 44 | DHFR | 6350315 | 5099 | 0.069 | 0.4755 | No | ||
| 45 | MCM10 | 4920632 | 5254 | 0.062 | 0.4681 | No | ||
| 46 | PSMB5 | 6290242 | 5566 | 0.050 | 0.4521 | No | ||
| 47 | CDKN1A | 4050088 6400706 | 6815 | 0.019 | 0.3851 | No | ||
| 48 | PSMD8 | 630142 | 7603 | 0.005 | 0.3427 | No | ||
| 49 | CCNA1 | 6290113 | 7823 | 0.001 | 0.3309 | No | ||
| 50 | PSMA6 | 50609 | 8309 | -0.006 | 0.3048 | No | ||
| 51 | POLA1 | 4760541 | 9007 | -0.018 | 0.2675 | No | ||
| 52 | POLE2 | 6110041 | 9182 | -0.021 | 0.2585 | No | ||
| 53 | PSMD5 | 3940139 4570041 | 9847 | -0.033 | 0.2231 | No | ||
| 54 | PSMC3 | 2480184 | 10249 | -0.041 | 0.2021 | No | ||
| 55 | PSMA3 | 5900047 7040161 | 10440 | -0.045 | 0.1925 | No | ||
| 56 | CDK7 | 2640451 | 10877 | -0.056 | 0.1698 | No | ||
| 57 | PSMB9 | 6980471 | 11460 | -0.072 | 0.1395 | No | ||
| 58 | PSMB4 | 520402 | 12769 | -0.127 | 0.0709 | No | ||
| 59 | MCM3 | 5570068 | 13403 | -0.172 | 0.0393 | No | ||
| 60 | POLA2 | 940519 | 13700 | -0.200 | 0.0263 | No | ||
| 61 | PSMA5 | 5390537 | 14212 | -0.258 | 0.0025 | No | ||
| 62 | PSMB3 | 4280594 | 14852 | -0.359 | -0.0266 | No | ||
| 63 | MCM5 | 2680647 | 15085 | -0.411 | -0.0330 | No | ||
| 64 | MCM2 | 5050139 | 15297 | -0.459 | -0.0376 | No | ||
| 65 | E2F1 | 5360093 | 15379 | -0.483 | -0.0348 | No | ||
| 66 | PSMB10 | 630504 6510039 | 15732 | -0.604 | -0.0449 | No | ||
| 67 | PSMB6 | 2690711 | 15755 | -0.612 | -0.0370 | No | ||
| 68 | PSMA7 | 2230131 | 16610 | -0.993 | -0.0683 | No | ||
| 69 | PSMD9 | 50020 5890368 | 17209 | -1.336 | -0.0808 | No | ||
| 70 | CCNE1 | 110064 | 17292 | -1.384 | -0.0647 | No | ||
| 71 | PSMC1 | 6350538 | 17333 | -1.414 | -0.0459 | No | ||
| 72 | PSMA1 | 380059 2760195 | 17467 | -1.500 | -0.0309 | No | ||
| 73 | PSMD2 | 4670706 5050364 | 17675 | -1.679 | -0.0171 | No | ||
| 74 | TK2 | 70725 | 18085 | -2.101 | -0.0081 | No | ||
| 75 | TYMS | 940450 1940068 3710008 5570546 | 18328 | -2.473 | 0.0155 | No |

