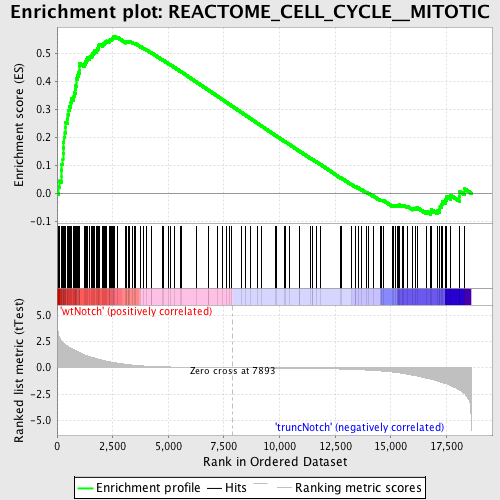
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | 0.5601966 |
| Normalized Enrichment Score (NES) | 1.7266458 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.16506091 |
| FWER p-Value | 0.382 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RFC4 | 3800082 6840142 | 64 | 3.081 | 0.0228 | Yes | ||
| 2 | PSMD14 | 5690593 | 119 | 2.806 | 0.0439 | Yes | ||
| 3 | NUP107 | 6380021 | 196 | 2.542 | 0.0615 | Yes | ||
| 4 | PSMA4 | 4560427 | 198 | 2.539 | 0.0831 | Yes | ||
| 5 | CCNB1 | 4590433 4780372 | 211 | 2.518 | 0.1039 | Yes | ||
| 6 | BUB1B | 1450288 | 261 | 2.413 | 0.1219 | Yes | ||
| 7 | POLE | 6020538 | 265 | 2.398 | 0.1422 | Yes | ||
| 8 | MAD2L1 | 4480725 | 273 | 2.379 | 0.1621 | Yes | ||
| 9 | PSMD7 | 2030619 6220594 | 293 | 2.337 | 0.1811 | Yes | ||
| 10 | CCNH | 3190347 | 309 | 2.289 | 0.1998 | Yes | ||
| 11 | PSMA2 | 6510093 | 353 | 2.215 | 0.2164 | Yes | ||
| 12 | RPA1 | 360452 | 385 | 2.159 | 0.2331 | Yes | ||
| 13 | RB1 | 5900338 | 390 | 2.152 | 0.2513 | Yes | ||
| 14 | FGFR1OP | 6510300 | 460 | 2.046 | 0.2650 | Yes | ||
| 15 | PSMD12 | 730044 | 488 | 2.007 | 0.2807 | Yes | ||
| 16 | PCNA | 940754 | 526 | 1.941 | 0.2953 | Yes | ||
| 17 | POLD3 | 6400278 | 553 | 1.899 | 0.3101 | Yes | ||
| 18 | MAPRE1 | 3290037 | 620 | 1.839 | 0.3222 | Yes | ||
| 19 | RFC1 | 1190673 | 630 | 1.828 | 0.3373 | Yes | ||
| 20 | SGOL2 | 2030338 | 755 | 1.693 | 0.3451 | Yes | ||
| 21 | CUL1 | 1990632 | 772 | 1.677 | 0.3585 | Yes | ||
| 22 | RPA3 | 5700136 | 821 | 1.631 | 0.3698 | Yes | ||
| 23 | MNAT1 | 730292 2350364 | 831 | 1.616 | 0.3832 | Yes | ||
| 24 | NDC80 | 4120465 | 867 | 1.583 | 0.3948 | Yes | ||
| 25 | YWHAG | 3780341 | 868 | 1.581 | 0.4083 | Yes | ||
| 26 | MCM4 | 2760673 5420711 | 904 | 1.538 | 0.4195 | Yes | ||
| 27 | ZWINT | 6940670 | 954 | 1.481 | 0.4295 | Yes | ||
| 28 | NUP160 | 1990551 | 1008 | 1.419 | 0.4388 | Yes | ||
| 29 | ORC5L | 1940133 1940711 | 1013 | 1.414 | 0.4506 | Yes | ||
| 30 | KIF23 | 5570112 | 1019 | 1.409 | 0.4624 | Yes | ||
| 31 | PSMD4 | 430068 | 1236 | 1.214 | 0.4611 | Yes | ||
| 32 | UBE2E1 | 4230064 1090440 | 1269 | 1.186 | 0.4694 | Yes | ||
| 33 | SMC1A | 3060600 5700148 5890113 6370154 | 1333 | 1.130 | 0.4757 | Yes | ||
| 34 | ZW10 | 2900735 3520687 | 1362 | 1.103 | 0.4836 | Yes | ||
| 35 | CETN2 | 3170148 | 1477 | 1.024 | 0.4862 | Yes | ||
| 36 | PRIM1 | 6420746 | 1539 | 0.999 | 0.4914 | Yes | ||
| 37 | CDC16 | 1940706 | 1589 | 0.980 | 0.4971 | Yes | ||
| 38 | KNTC1 | 430079 | 1652 | 0.940 | 0.5018 | Yes | ||
| 39 | GMNN | 2630148 | 1691 | 0.916 | 0.5075 | Yes | ||
| 40 | RAD21 | 1990278 | 1781 | 0.856 | 0.5100 | Yes | ||
| 41 | NUP43 | 3360482 | 1810 | 0.839 | 0.5157 | Yes | ||
| 42 | PSMD3 | 1400647 | 1854 | 0.811 | 0.5203 | Yes | ||
| 43 | CENPA | 5080154 | 1872 | 0.803 | 0.5262 | Yes | ||
| 44 | BIRC5 | 110408 580014 1770632 | 1894 | 0.790 | 0.5318 | Yes | ||
| 45 | CDC45L | 70537 3130114 | 2050 | 0.690 | 0.5293 | Yes | ||
| 46 | CSNK1E | 2850347 5050093 6110301 | 2070 | 0.684 | 0.5341 | Yes | ||
| 47 | PSMC5 | 2760315 6550021 | 2143 | 0.650 | 0.5358 | Yes | ||
| 48 | CCNA2 | 5290075 | 2176 | 0.634 | 0.5395 | Yes | ||
| 49 | CDC25A | 3800184 | 2212 | 0.617 | 0.5428 | Yes | ||
| 50 | MCM7 | 3290292 5220056 | 2334 | 0.569 | 0.5411 | Yes | ||
| 51 | PSMC6 | 2810021 | 2356 | 0.560 | 0.5448 | Yes | ||
| 52 | SEC13 | 380577 | 2387 | 0.549 | 0.5478 | Yes | ||
| 53 | PSMC4 | 580050 1580025 | 2463 | 0.518 | 0.5482 | Yes | ||
| 54 | PSME3 | 2810537 | 2475 | 0.515 | 0.5520 | Yes | ||
| 55 | PRKAR2B | 3130593 5220577 | 2512 | 0.501 | 0.5543 | Yes | ||
| 56 | ORC4L | 4230538 5550288 | 2535 | 0.494 | 0.5574 | Yes | ||
| 57 | STAG2 | 4540132 | 2560 | 0.485 | 0.5602 | Yes | ||
| 58 | RANBP2 | 4280338 | 2707 | 0.437 | 0.5560 | No | ||
| 59 | PSMB1 | 2940402 | 3086 | 0.317 | 0.5383 | No | ||
| 60 | PSMB2 | 940035 4210324 | 3097 | 0.313 | 0.5404 | No | ||
| 61 | FEN1 | 1770541 | 3136 | 0.302 | 0.5409 | No | ||
| 62 | CDK2 | 130484 2260301 4010088 5050110 | 3216 | 0.284 | 0.5391 | No | ||
| 63 | RPA2 | 2760301 5420195 | 3221 | 0.283 | 0.5413 | No | ||
| 64 | TUBG1 | 3140204 | 3242 | 0.280 | 0.5426 | No | ||
| 65 | PPP1CC | 6380300 2510647 | 3376 | 0.251 | 0.5375 | No | ||
| 66 | ORC1L | 2370328 6110390 | 3473 | 0.230 | 0.5343 | No | ||
| 67 | CLASP1 | 6860279 | 3529 | 0.218 | 0.5332 | No | ||
| 68 | YWHAE | 5310435 | 3768 | 0.178 | 0.5218 | No | ||
| 69 | WEE1 | 3390070 | 3884 | 0.160 | 0.5169 | No | ||
| 70 | ORC2L | 1990470 6510019 | 4029 | 0.144 | 0.5104 | No | ||
| 71 | CDC25C | 2570673 4760161 6520707 | 4241 | 0.124 | 0.5000 | No | ||
| 72 | OFD1 | 4810670 | 4249 | 0.123 | 0.5007 | No | ||
| 73 | PTTG1 | 520463 | 4733 | 0.086 | 0.4753 | No | ||
| 74 | DCTN3 | 6840131 | 4767 | 0.085 | 0.4742 | No | ||
| 75 | PSMD11 | 2340538 6510053 | 5028 | 0.072 | 0.4608 | No | ||
| 76 | DHFR | 6350315 | 5099 | 0.069 | 0.4576 | No | ||
| 77 | MCM10 | 4920632 | 5254 | 0.062 | 0.4497 | No | ||
| 78 | PSMB5 | 6290242 | 5566 | 0.050 | 0.4333 | No | ||
| 79 | CCNB2 | 6510528 | 5570 | 0.050 | 0.4336 | No | ||
| 80 | SDCCAG8 | 1780687 | 6284 | 0.030 | 0.3953 | No | ||
| 81 | CDKN1A | 4050088 6400706 | 6815 | 0.019 | 0.3667 | No | ||
| 82 | CENPC1 | 610273 | 6826 | 0.018 | 0.3664 | No | ||
| 83 | CLASP2 | 2510139 | 7204 | 0.011 | 0.3460 | No | ||
| 84 | UBE2D1 | 1850400 | 7452 | 0.007 | 0.3327 | No | ||
| 85 | PSMD8 | 630142 | 7603 | 0.005 | 0.3247 | No | ||
| 86 | XPO1 | 540707 | 7753 | 0.002 | 0.3166 | No | ||
| 87 | CCNA1 | 6290113 | 7823 | 0.001 | 0.3129 | No | ||
| 88 | PSMA6 | 50609 | 8309 | -0.006 | 0.2867 | No | ||
| 89 | BUB1 | 5390270 | 8457 | -0.009 | 0.2788 | No | ||
| 90 | DYNLL1 | 5570113 | 8670 | -0.013 | 0.2674 | No | ||
| 91 | POLA1 | 4760541 | 9007 | -0.018 | 0.2494 | No | ||
| 92 | POLE2 | 6110041 | 9182 | -0.021 | 0.2402 | No | ||
| 93 | KIF2A | 3990286 6130575 | 9800 | -0.032 | 0.2071 | No | ||
| 94 | PSMD5 | 3940139 4570041 | 9847 | -0.033 | 0.2048 | No | ||
| 95 | SGOL1 | 1980075 5220092 6020711 | 10214 | -0.040 | 0.1854 | No | ||
| 96 | PSMC3 | 2480184 | 10249 | -0.041 | 0.1839 | No | ||
| 97 | PSMA3 | 5900047 7040161 | 10440 | -0.045 | 0.1740 | No | ||
| 98 | CDK7 | 2640451 | 10877 | -0.056 | 0.1509 | No | ||
| 99 | NEK2 | 2120520 | 10893 | -0.056 | 0.1505 | No | ||
| 100 | PAFAH1B1 | 4230333 6420121 6450066 | 11397 | -0.070 | 0.1239 | No | ||
| 101 | PSMB9 | 6980471 | 11460 | -0.072 | 0.1211 | No | ||
| 102 | RFC3 | 1980600 | 11471 | -0.072 | 0.1212 | No | ||
| 103 | NUP37 | 2370097 6370435 6380008 | 11643 | -0.077 | 0.1126 | No | ||
| 104 | CLIP1 | 2850162 6770397 | 11827 | -0.084 | 0.1034 | No | ||
| 105 | CENPE | 2850022 | 12747 | -0.126 | 0.0548 | No | ||
| 106 | PSMB4 | 520402 | 12769 | -0.127 | 0.0547 | No | ||
| 107 | PLK1 | 1780369 2640121 | 13227 | -0.159 | 0.0313 | No | ||
| 108 | MCM3 | 5570068 | 13403 | -0.172 | 0.0233 | No | ||
| 109 | UBE2C | 6130017 | 13412 | -0.173 | 0.0244 | No | ||
| 110 | KIF20A | 2650050 | 13526 | -0.183 | 0.0198 | No | ||
| 111 | POLA2 | 940519 | 13700 | -0.200 | 0.0122 | No | ||
| 112 | TUBA1A | 610128 4540347 6420070 6980181 | 13900 | -0.220 | 0.0033 | No | ||
| 113 | PLK4 | 430162 5720110 | 14001 | -0.232 | -0.0002 | No | ||
| 114 | PSMA5 | 5390537 | 14212 | -0.258 | -0.0093 | No | ||
| 115 | ACTR1A | 5860070 | 14546 | -0.308 | -0.0247 | No | ||
| 116 | SSNA1 | 360593 | 14597 | -0.317 | -0.0247 | No | ||
| 117 | DCTN2 | 540471 3780717 | 14688 | -0.333 | -0.0267 | No | ||
| 118 | MCM5 | 2680647 | 15085 | -0.411 | -0.0447 | No | ||
| 119 | CDC20 | 3440017 3440044 6220088 | 15103 | -0.413 | -0.0421 | No | ||
| 120 | BUB3 | 3170546 | 15220 | -0.440 | -0.0446 | No | ||
| 121 | MCM2 | 5050139 | 15297 | -0.459 | -0.0448 | No | ||
| 122 | POLD2 | 6400148 | 15333 | -0.469 | -0.0427 | No | ||
| 123 | E2F1 | 5360093 | 15379 | -0.483 | -0.0410 | No | ||
| 124 | TUBA4A | 6900279 | 15519 | -0.530 | -0.0440 | No | ||
| 125 | NUMA1 | 6520576 | 15578 | -0.548 | -0.0425 | No | ||
| 126 | PSMB10 | 630504 6510039 | 15732 | -0.604 | -0.0456 | No | ||
| 127 | PRKACA | 2640731 4050048 | 15991 | -0.708 | -0.0535 | No | ||
| 128 | LIG1 | 1980438 4540112 5570609 | 16090 | -0.754 | -0.0524 | No | ||
| 129 | TUBB4 | 5310681 | 16184 | -0.789 | -0.0507 | No | ||
| 130 | PSMA7 | 2230131 | 16610 | -0.993 | -0.0652 | No | ||
| 131 | RANGAP1 | 2320593 6650601 | 16802 | -1.071 | -0.0664 | No | ||
| 132 | POLD1 | 4830026 | 16821 | -1.079 | -0.0581 | No | ||
| 133 | RFC5 | 3800452 6020091 | 17085 | -1.251 | -0.0617 | No | ||
| 134 | PMF1 | 5130273 | 17191 | -1.325 | -0.0561 | No | ||
| 135 | PSMD9 | 50020 5890368 | 17209 | -1.336 | -0.0456 | No | ||
| 136 | CCNE1 | 110064 | 17292 | -1.384 | -0.0382 | No | ||
| 137 | PSMC1 | 6350538 | 17333 | -1.414 | -0.0283 | No | ||
| 138 | PSMA1 | 380059 2760195 | 17467 | -1.500 | -0.0227 | No | ||
| 139 | CDC25B | 6940102 | 17519 | -1.538 | -0.0123 | No | ||
| 140 | PSMD2 | 4670706 5050364 | 17675 | -1.679 | -0.0064 | No | ||
| 141 | TK2 | 70725 | 18085 | -2.101 | -0.0106 | No | ||
| 142 | CDK4 | 540075 4540600 | 18101 | -2.120 | 0.0067 | No | ||
| 143 | TYMS | 940450 1940068 3710008 5570546 | 18328 | -2.473 | 0.0156 | No |

