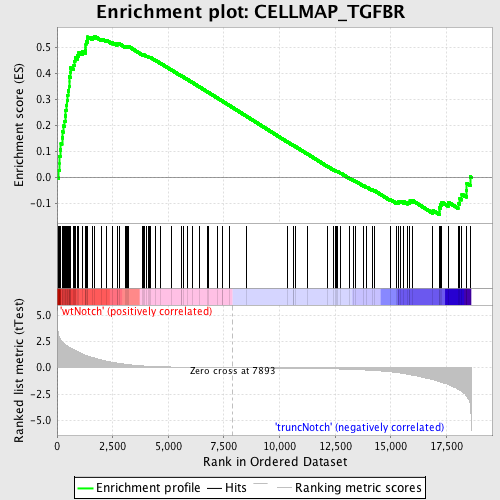
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_truncNotch_versus_wtNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch.phenotype_truncNotch_versus_wtNotch.cls #wtNotch_versus_truncNotch_repos |
| Phenotype | phenotype_truncNotch_versus_wtNotch.cls#wtNotch_versus_truncNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | CELLMAP_TGFBR |
| Enrichment Score (ES) | 0.5438832 |
| Normalized Enrichment Score (NES) | 1.636012 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.20721008 |
| FWER p-Value | 0.861 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SMAD4 | 5670519 | 68 | 3.059 | 0.0267 | Yes | ||
| 2 | ANAPC1 | 6350162 | 106 | 2.840 | 0.0529 | Yes | ||
| 3 | KPNB1 | 1690138 | 110 | 2.828 | 0.0808 | Yes | ||
| 4 | NFYB | 1850053 | 138 | 2.739 | 0.1065 | Yes | ||
| 5 | CTCF | 5340017 | 172 | 2.618 | 0.1307 | Yes | ||
| 6 | RBL1 | 3130372 | 220 | 2.502 | 0.1530 | Yes | ||
| 7 | STRAP | 1340170 | 252 | 2.428 | 0.1754 | Yes | ||
| 8 | ANAPC4 | 4540338 | 272 | 2.384 | 0.1981 | Yes | ||
| 9 | CCND1 | 460524 770309 3120576 6980398 | 346 | 2.224 | 0.2162 | Yes | ||
| 10 | ZEB1 | 6840162 | 367 | 2.192 | 0.2369 | Yes | ||
| 11 | RB1 | 5900338 | 390 | 2.152 | 0.2571 | Yes | ||
| 12 | FNTA | 70059 | 409 | 2.120 | 0.2771 | Yes | ||
| 13 | SNX2 | 2470068 3360364 6510064 | 444 | 2.069 | 0.2958 | Yes | ||
| 14 | SNX4 | 1980358 | 469 | 2.037 | 0.3148 | Yes | ||
| 15 | UBE2L3 | 1690707 5900242 | 491 | 2.003 | 0.3335 | Yes | ||
| 16 | MAP3K7 | 6040068 | 534 | 1.929 | 0.3504 | Yes | ||
| 17 | UBE2D3 | 3190452 | 542 | 1.915 | 0.3690 | Yes | ||
| 18 | NCOA1 | 3610438 | 543 | 1.911 | 0.3880 | Yes | ||
| 19 | ANAPC5 | 730164 7100685 | 597 | 1.862 | 0.4036 | Yes | ||
| 20 | PIAS1 | 5340301 | 607 | 1.854 | 0.4215 | Yes | ||
| 21 | COPS5 | 1340722 5050215 | 751 | 1.699 | 0.4307 | Yes | ||
| 22 | CUL1 | 1990632 | 772 | 1.677 | 0.4462 | Yes | ||
| 23 | SMAD2 | 4200592 | 806 | 1.643 | 0.4608 | Yes | ||
| 24 | PRKCD | 770592 | 914 | 1.525 | 0.4701 | Yes | ||
| 25 | HDAC1 | 2850670 | 976 | 1.456 | 0.4813 | Yes | ||
| 26 | NFYA | 5860368 | 1139 | 1.293 | 0.4854 | Yes | ||
| 27 | UBE2E1 | 4230064 1090440 | 1269 | 1.186 | 0.4902 | Yes | ||
| 28 | SP1 | 6590017 | 1291 | 1.160 | 0.5005 | Yes | ||
| 29 | MAP3K7IP1 | 2120095 5890452 6040167 | 1294 | 1.155 | 0.5119 | Yes | ||
| 30 | MAPK14 | 5290731 | 1301 | 1.150 | 0.5230 | Yes | ||
| 31 | UBE2D2 | 4590671 6510446 | 1363 | 1.102 | 0.5306 | Yes | ||
| 32 | CREBBP | 5690035 7040050 | 1378 | 1.091 | 0.5407 | Yes | ||
| 33 | CDC16 | 1940706 | 1589 | 0.980 | 0.5391 | Yes | ||
| 34 | TFDP1 | 1980112 | 1671 | 0.926 | 0.5439 | Yes | ||
| 35 | MAPK8 | 2640195 | 2013 | 0.718 | 0.5326 | No | ||
| 36 | CDC25A | 3800184 | 2212 | 0.617 | 0.5280 | No | ||
| 37 | PRKAR2A | 2340136 | 2483 | 0.512 | 0.5185 | No | ||
| 38 | SNW1 | 4010736 | 2694 | 0.442 | 0.5115 | No | ||
| 39 | TGFB3 | 1070041 | 2702 | 0.438 | 0.5155 | No | ||
| 40 | ATF2 | 2260348 2480441 | 2797 | 0.409 | 0.5145 | No | ||
| 41 | AR | 6380167 | 3059 | 0.323 | 0.5036 | No | ||
| 42 | CAMK2B | 2760041 | 3126 | 0.306 | 0.5031 | No | ||
| 43 | AXIN1 | 3360358 6940451 | 3148 | 0.299 | 0.5049 | No | ||
| 44 | CDK2 | 130484 2260301 4010088 5050110 | 3216 | 0.284 | 0.5041 | No | ||
| 45 | SNX1 | 3190670 | 3837 | 0.169 | 0.4723 | No | ||
| 46 | RUNX2 | 1400193 4230215 | 3873 | 0.162 | 0.4720 | No | ||
| 47 | TGFBR2 | 1780711 1980537 6550398 | 3914 | 0.157 | 0.4714 | No | ||
| 48 | MAP2K6 | 1230056 2940204 | 4023 | 0.145 | 0.4670 | No | ||
| 49 | HOXA9 | 610494 4730040 5130601 | 4121 | 0.134 | 0.4631 | No | ||
| 50 | RBX1 | 2120010 2340047 | 4144 | 0.132 | 0.4632 | No | ||
| 51 | RBL2 | 580446 1400670 | 4187 | 0.128 | 0.4622 | No | ||
| 52 | TGFBR1 | 1400148 4280020 6550711 | 4403 | 0.109 | 0.4517 | No | ||
| 53 | EID2 | 3130176 | 4648 | 0.092 | 0.4394 | No | ||
| 54 | CDC23 | 3190593 | 5157 | 0.066 | 0.4126 | No | ||
| 55 | CCNB2 | 6510528 | 5570 | 0.050 | 0.3909 | No | ||
| 56 | FOSB | 1940142 | 5598 | 0.049 | 0.3899 | No | ||
| 57 | NFYC | 1400746 | 5660 | 0.047 | 0.3871 | No | ||
| 58 | TGFBR3 | 5290577 | 5843 | 0.041 | 0.3776 | No | ||
| 59 | NUP153 | 7000452 | 6085 | 0.035 | 0.3650 | No | ||
| 60 | MYC | 380541 4670170 | 6382 | 0.028 | 0.3493 | No | ||
| 61 | ANAPC10 | 870086 1170037 2260129 | 6773 | 0.019 | 0.3284 | No | ||
| 62 | CDKN1A | 4050088 6400706 | 6815 | 0.019 | 0.3263 | No | ||
| 63 | SKIL | 1090364 | 7196 | 0.012 | 0.3059 | No | ||
| 64 | SMAD7 | 430377 | 7413 | 0.008 | 0.2943 | No | ||
| 65 | UBE2D1 | 1850400 | 7452 | 0.007 | 0.2924 | No | ||
| 66 | XPO1 | 540707 | 7753 | 0.002 | 0.2762 | No | ||
| 67 | ESR1 | 4060372 5860193 | 8512 | -0.010 | 0.2353 | No | ||
| 68 | ZFYVE9 | 6980288 7100017 | 10363 | -0.043 | 0.1358 | No | ||
| 69 | CITED1 | 2350670 | 10624 | -0.050 | 0.1223 | No | ||
| 70 | CAV1 | 870025 | 10632 | -0.050 | 0.1224 | No | ||
| 71 | PRKAR1B | 6130411 | 10735 | -0.052 | 0.1174 | No | ||
| 72 | SMAD6 | 870504 | 11267 | -0.066 | 0.0893 | No | ||
| 73 | ATF3 | 1940546 | 12166 | -0.097 | 0.0418 | No | ||
| 74 | ENG | 4280270 | 12416 | -0.109 | 0.0294 | No | ||
| 75 | JUN | 840170 | 12506 | -0.114 | 0.0257 | No | ||
| 76 | ANAPC2 | 6660438 | 12550 | -0.116 | 0.0246 | No | ||
| 77 | YAP1 | 4230577 | 12593 | -0.118 | 0.0235 | No | ||
| 78 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 12759 | -0.127 | 0.0158 | No | ||
| 79 | SDC2 | 1980168 | 13129 | -0.151 | -0.0026 | No | ||
| 80 | SNX6 | 6200086 | 13326 | -0.166 | -0.0116 | No | ||
| 81 | SMURF2 | 1940161 | 13405 | -0.172 | -0.0141 | No | ||
| 82 | HSPA8 | 2120315 1050132 6550746 | 13775 | -0.208 | -0.0319 | No | ||
| 83 | FOXO3 | 2510484 4480451 | 13919 | -0.222 | -0.0374 | No | ||
| 84 | ZEB2 | 4120070 4150215 | 14153 | -0.250 | -0.0476 | No | ||
| 85 | MEF2A | 2350041 3360128 6040180 | 14275 | -0.266 | -0.0514 | No | ||
| 86 | FOS | 1850315 | 14971 | -0.386 | -0.0852 | No | ||
| 87 | TP53 | 6130707 | 15236 | -0.442 | -0.0950 | No | ||
| 88 | CDK6 | 4920253 | 15335 | -0.469 | -0.0957 | No | ||
| 89 | ETS1 | 5270278 6450717 6620465 | 15339 | -0.470 | -0.0912 | No | ||
| 90 | JUNB | 4230048 | 15452 | -0.504 | -0.0922 | No | ||
| 91 | DAB2 | 60309 | 15583 | -0.551 | -0.0938 | No | ||
| 92 | TGFB1 | 1940162 | 15749 | -0.610 | -0.0966 | No | ||
| 93 | VDR | 130156 510438 | 15833 | -0.637 | -0.0948 | No | ||
| 94 | BTRC | 3170131 | 15860 | -0.646 | -0.0898 | No | ||
| 95 | CAMK2G | 630097 | 15963 | -0.695 | -0.0884 | No | ||
| 96 | FKBP1A | 2450368 | 16894 | -1.130 | -0.1274 | No | ||
| 97 | FZR1 | 6220504 6660670 | 17180 | -1.320 | -0.1297 | No | ||
| 98 | STK11IP | 6980735 | 17182 | -1.321 | -0.1167 | No | ||
| 99 | DAXX | 4010647 | 17225 | -1.342 | -0.1056 | No | ||
| 100 | CCNE1 | 110064 | 17292 | -1.384 | -0.0954 | No | ||
| 101 | SNIP1 | 5360092 | 17577 | -1.585 | -0.0950 | No | ||
| 102 | PIK3R2 | 1660193 | 18023 | -2.025 | -0.0990 | No | ||
| 103 | CDK4 | 540075 4540600 | 18101 | -2.120 | -0.0821 | No | ||
| 104 | SMAD3 | 6450671 | 18195 | -2.238 | -0.0649 | No | ||
| 105 | ANAPC7 | 5260274 | 18397 | -2.652 | -0.0494 | No | ||
| 106 | CAMK2D | 2370685 | 18418 | -2.728 | -0.0234 | No | ||
| 107 | ARRB2 | 2060441 | 18568 | -3.434 | 0.0026 | No |

